Abstract
The reduction of iron is an essential step in the transferrin (Tf) cycle, which is the dominant pathway for iron uptake by red blood cell (RBC) precursors. A deficiency in RBC iron acquisition leads to a hypochromic, microcytic anemia. Using a positional cloning strategy, we have identified a gene, six-transmembrane epithelial antigen of the prostate 3 (Steap3), which is responsible for the iron deficiency anemia in the murine mutant nm1054. Steap3 is expressed highly in hematopoietic tissues, co-localizes with the Tf cycle endosome, and facilitates Tf-bound iron uptake. Steap3 shares homology with F420H2:NADP+ oxidoreductases found in archaea and bacteria, as well as with the yeast FRE family of metalloreductases. Overexpression of Steap3 stimulates the reduction of iron, and mice lacking Steap3 are deficient in erythroid ferrireductase activity. Altogether, these findings demonstrate that Steap3 is an endosomal ferrireductase required for efficient Tf-dependent iron uptake in erythroid cells.
Red blood cell precursors are uniquely dependent upon the transferrin (Tf) cycle to acquire iron in order to synthesize heme in amounts sufficient for hemoglobin production1. In the transferrin cycle, iron bound to transferrin (Tf) binds to the transferrin receptor (Tfr1), the complex is taken up by receptor-mediated endocytosis, and iron is released from Tf by endosomal acidification to be delivered to the cytoplasm by the divalent metal transporter 1 (Dmt1)2–4. Tf carries ferric iron (Fe3+), whereas Dmt1 is selective for ferrous iron (Fe2+)4. Therefore, iron must be reduced in the Tf cycle endosome. Despite functional evidence of such an activity, the molecular identity of this reductase is unknown. Recently, an ascorbate-dependent b-type cytochrome ferrireductase, Dcytb (Cybrd1), expressed predominantly in the duodenum, was described5. However, Dcytb is not expressed highly in erythroid precursors, and Dcytb null mice have normal iron metabolism and normal hematologic parameters6. Such data argue against a significant role for Dcytb in the Tf cycle. Alternatively, several studies have demonstrated the existence of an NAD(P)H-dependent, cell-surface and/or endosomal ferrireductase activity in many cell types, including erythroid cells7–9. We now demonstrate that the major erythroid ferrireductase of the Tf cycle endosome is Steap3, which is predicted to be a flavin-NADPH-dependent, membrane-bound oxidoreductase.
Previously, we described the phenotype of a murine mutant, nm1054, which has a microcytic, hypochromic anemia10. In order to identify the gene responsible for the anemia, we took a positional cloning approach. We mapped nm1054 (nm) to a 0.5 centimorgan (cM) interval on mouse chromosome 1 in 542 affected [CBA/J-nm1054 x CAST/Ei] F2 intercross animals10, and defined a minimal physical interval for the locus between microsatellite markers D1Mit27 and D1Mit471 (Supplementary Fig. 1 online). Three contiguous markers, D1Mit54, D1Mit191, and D1Mit389, could not be amplified in affected animals, suggesting that the nm1054 mutation was a large genomic deletion. Serial analysis of genomic PCR amplicons across the region confirmed this observation, and defined a deletion of approximately 400 kb that contained all or part of 6 genes (Fig. 1a). RT-PCR, southern blot, and/or northern blot analysis demonstrated that each of these genes was absent in nm/nm animals (not shown). A BAC contig of the deletion was developed, and transgenic lines were created from four overlapping BAC clones that spanned the entire deletion without disrupting any of the genes (Fig. 1a). Two transgenic lines derived from RPCI-22 BAC clone 11D19, which contains 2 genes, corrected the anemia (Fig. 1b). The first, located within the deletion interval, is six-transmembrane epithelial antigen of the prostate 3 (Steap3, also known as pHyde11–13, TSAP614,15, and Dudulin2 [GenBank AY029586.1]). The second, which lies outside the deletion, is complement component 1, q subcomponent 2-like (C1ql2). Two partial BAC 11D19 insertions containing C1ql2 alone, failed to complement the anemia, suggesting that Steap3 was the anemia gene on BAC 11D19.
Figure 1.
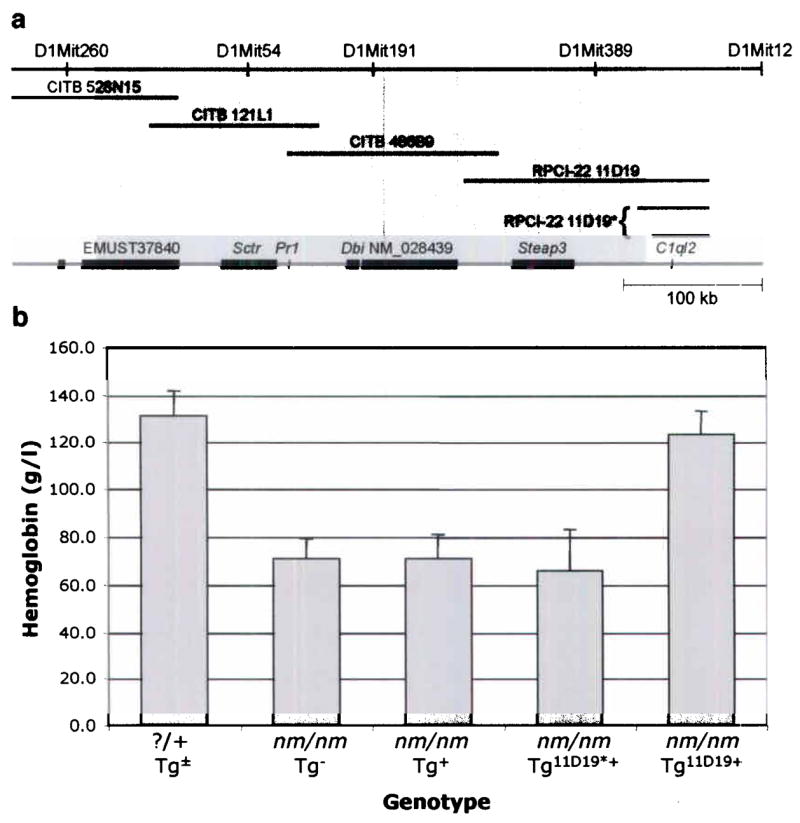
nm1054 physical map and BAC complementation. (a) Physical map of the nm1054 region demonstrating the extent of the deletion interval (shaded region) based on sequences in the Ensembl database (www.ensembl.org; April 2005 release). Microsatellite markers (top), BAC transgenes (middle), and genes (bottom) are shown. CITB and RPCI-22 BAC transgenes are indicated by their clone ID numbers. Partial insertions of RPCI-22 11D19 are indicated by an asterix (*). Ensembl transcript ENSMUST00000037840; Sctr, secretin receptor; Pr1, Neuronal voltage-gated calcium channel γ-like subunit; Dbi, diazepam binding inhibitor; RefSeq_dna ID NM_028439; Steap3, six-transmembrane epithelial antigen of the prostate 3; C1ql2, complement component 1, q subcomponent 2-like (b) Comparison of hemoglobin (HGB [g/l]) levels in 4 week old BAC transgenic [C57BL/6 x 129S6/SvEvTac] F1 animals. ?/+ includes all nm/+ and +/+ animals with or without a BAC transgene (n=12). Other groups are nm/nm animals without a BAC transgene (Tg−; n=11), a transgene other than RPCI-22 11D19 (Tg+; n=15), a partial insertion of RPCI-22 11D19 (Tg11D19*+; n=6), or a complete insertion of RPCI-22 11D19 (Tg11D19+). Error bars ± 1 standard deviation (S.D.).
Because the nm1054 anemia is intrinsic to the hematopoietic compartment and manifests as a Tf-dependent iron uptake defect10, we expected the anemia gene to be expressed in hematopoietic tissues, and the protein to be present in a physiologically relevant cellular location. In situ hybridization analysis of mouse embryos and adult tissues, as well as quantitative real-time PCR of human tissues, demonstrated that Steap3 is highly expressed in fetal liver (the site of mid-gestational hematopoiesis), adult bone marrow, placenta, liver, skeletal muscle, and pancreas (Fig. 2a,b). This pattern of distribution supports a role for Steap3 in iron metabolism, particularly in cells, such as erythroid precursors, that are highly dependent upon Tf for iron acquisition. Furthermore, we demonstrated that a stably expressed, epitope-tagged form of Steap3 partially co-localizes with Tf, Tfr1, and Dmt1 within endosomes (Fig. 3a–i). Also similar to Tfr1 and Dmt1, in transiently transfected cells expressing high levels of Steap3, the protein could be detected on the cell surface by immunofluorescence (not shown). This co-localization with three other key components of Tf-dependent iron uptake, places Steap3 in a sub-cellular compartment equipped for high-affinity, high avidity iron transport.
Figure 2.
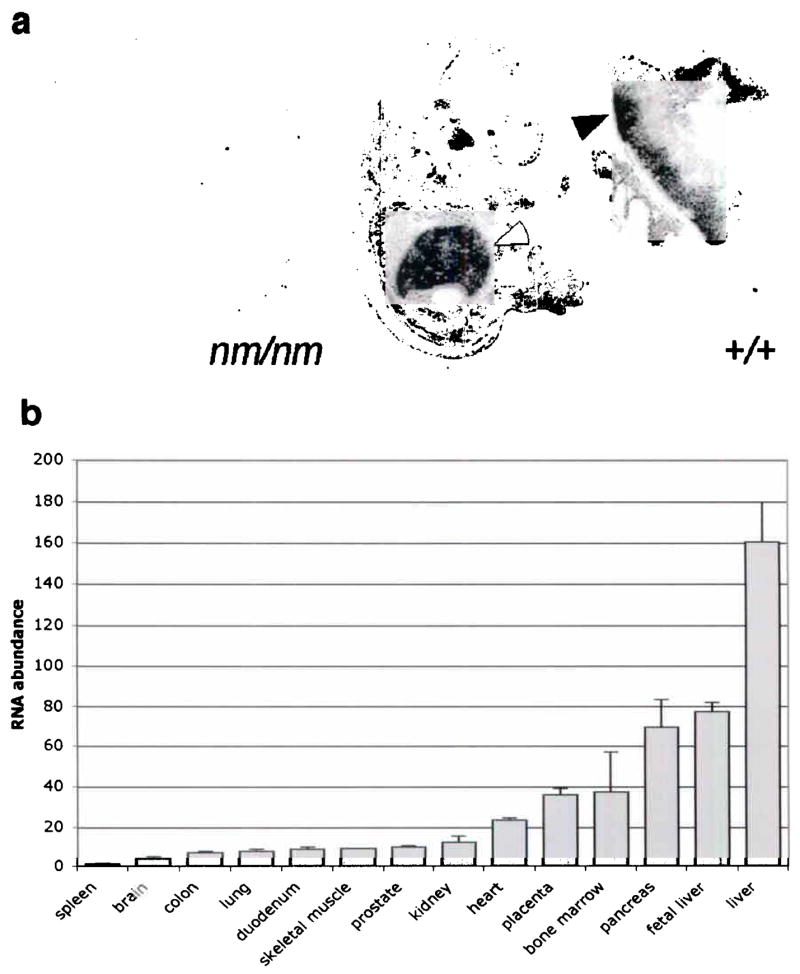
Steap3 mRNA expression (a) In situ hybridization of E15.5 mouse embryo demonstrating high-level fetal liver (open arrow) and labyrinthine placental expression (closed arrow). (b) Quantitative real time PCR of STEAP3 in human tissues. Relative RNA abundance is normalized to spleen, which was defined as a ratio of 1.0. Error bars ± 1 S.D.
Figure 3.
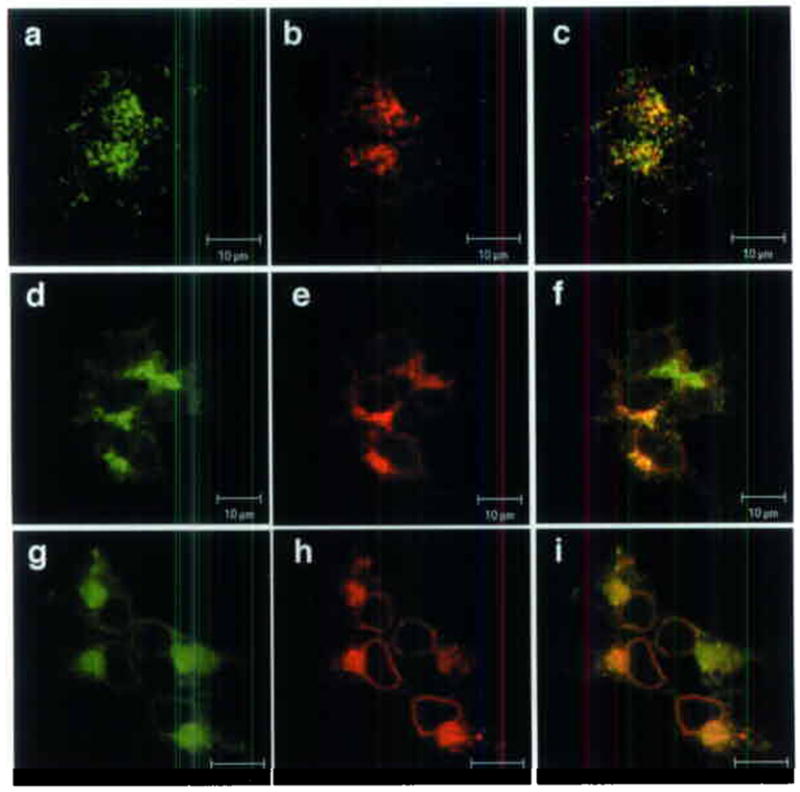
Steap3 sub-cellular localization. (a–i) Co-localization of epitope-tagged Steap3 with endogenous Tf and Tfr1, and epitope-tagged DMT1. (a) Steap3, (b) Tf, (c) Tf-Steap3 merged; (d) Steap3, (e) Tfr1, (f) Tfr1-Steap3 merged, (g) Steap3, (h) Dmt1, (i) Dmt1-Steap3 merged.
To further assess if Steap3 was the nm1054 anemia gene, we attempted to somatically correct the hematopoietic defect in vivo. Mutant fetal liver hematopoietic cells were retrovirally transduced with a wild-type copy of Steap3 and transplanted into lethally irradiated wild type mice. Expression of Steap3 rescued the anemia, as indicated by an increase in the mean cell volume (MCV) and cellular hemoglobin content of the reticulocytes (CHr). Furthermore, because the proportion of protoporphyrin IX complexed with zinc (ZnPP) increases in states of iron deficiency16, the decreased ZnPP/Heme in the complemented mutant reflects specific correction of the intraerythroid iron deficiency (Fig. 4a–c). In total, these data demonstrate Steap3 rescues the hematopoietic defect in nm1054.
Figure 4.
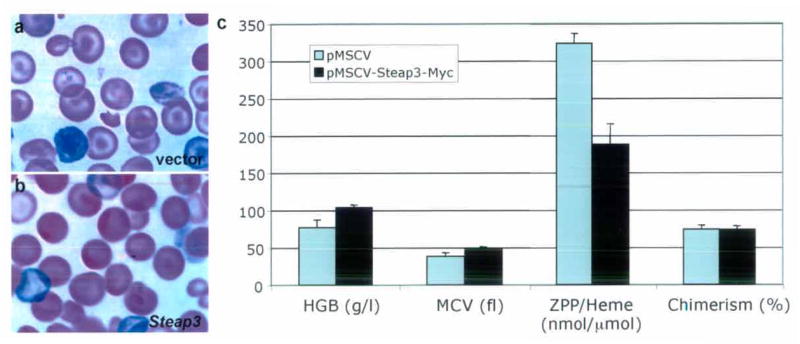
Somatic complementation of the nm1054 anemia in bone marrow chimeras. (a,b) Comparison of peripheral blood smears of lethally irradiated wild type recipient mice transplanted with homozygous mutant fetal liver hematopoietic cells transduced with (a) the retroviral vector alone, or (b) a construct expressing Steap3. (c) Hematological data and RBC chimerism in transplant chimeras at 4 weeks post-transplantation (n=4 in each group). HGB=hemoglobin, MCV=mean RBC volume, ZPP/Heme=RBC zinc protoporphyrin IX: iron protoporphyrin IX (heme) ratio. Error bars ± 1 S.D. Student’s T-test P= 0.009 (HGB), 0.017 (MCV), <0.001 (ZPP/Heme). Chimerism was not significantly different between the two groups (P=0.837). Error bars ± 1 S.D.
To further expand upon the Steap3 complementation result, we created a Steap3 targeted deletion allele (Supplementary Figure 2 online). Phenotypic analysis of [129S4/SvJae-Steap3+/− x 129S6/SvEvTac-nm1054/+] F2 intercross progeny demonstrated that the nm1054 anemia was allelic with the Steap3 targeted deletion, and that homozygosity for the Steap3 null allele recapitulated the homozygous nm1054 anemia phenotype (Fig. 5a–e and Supplementary Table 1 and Supplementary Table 2 online). Furthermore, Steap3−/− reticulocytes, like nm/nm reticulocytes10, have a defect in Tf-dependent iron uptake. Steap3−/− reticulocytes are more than 3 fold less efficient in acquiring iron from Tf when compared to wild type littermate controls (Fig. 5f). Upon re-incubation with media containing an excess of apo-Tf, they also lose a substantial fraction of iron initially taken up into the cell (17.8±3.3%), whereas control cells lose virtually none at all (−1.6±3.7%). Taken together, these data conclusively demonstrate that the nm1054 anemia is due to loss of Steap3, that nm1054 is a deletion allele of Steap3 (Steap3nm1054), and that mutation of Steap3 results in a defect in Tf-mediated iron uptake.
Figure 5.
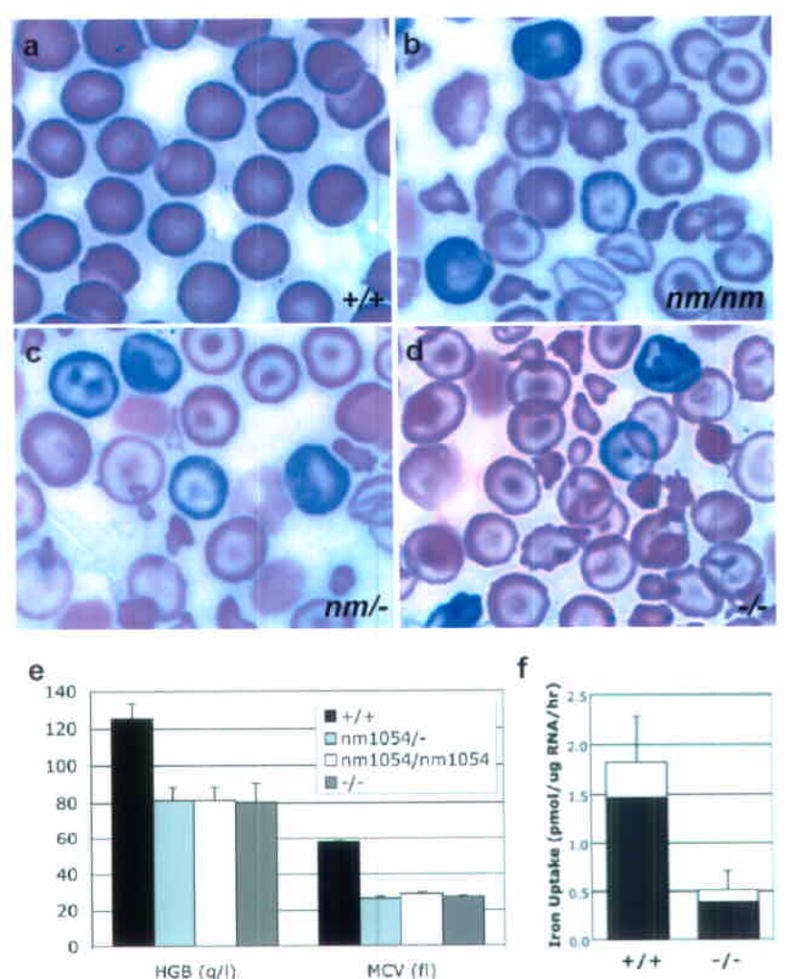
Hematologic data from nm1054 and Steap3 null mice. (a–d) Peripheral blood smears of (a) Steap3+/+, (b) Steap3nm1054/nm1054, (c) Steap3nm1054/−, and (d) Steap3−/− animals at 8 weeks of age. (e) Hematological data in 8 week old Steap3+/+ (n=5), Steap3nm1054/nm1054 (n=6), Steap3nm/1054/− (n=6), and Steap3−/− (n=6) mice. Error bars ± 1 S.D. See also Supplementary Table 1 online. (f) Iron uptake activity in Steap3+/+ (n=5) and Steap3−/− (n=6) reticulocyte-rich RBCs. The fraction of the total iron, present as heme, is indicated in the filled portion of the bar.
Sequence analysis predicts Steap3 to have an N-terminal oxidoreductase domain and a C-terminal transmembrane region (Fig. 6a). The oxidoreductase domain is predicted to be an unusual flavin-NAD(P)H binding structure (PFAM 03807.8)17 homologous to one found in archaea and bacteria that utilizes NADPH and the modified flavin F420 as cofactors18,19. This domain is not present in yeast, C. elegans, or Drosophila, and only two other structurally similar mammalian proteins, Steap4 (Tiarp or Tnfaip920) and Steap221 (STAMP1), have homologous domains (Fig. 6b). The C-terminal region of the protein is predicted to have six transmembrane helices that are very distantly related to the S. cerevisiae FRE family of b-type cytochrome metalloreductases, which are essential for iron and copper uptake18,22 (Fig. 6b). In particular, two of four histidine residues implicated in binding heme in the FRE proteins23 are conserved in all four Steap family members, including Steap24,25, which lacks the N-terminal oxidoreductase domain (Fig. 6b). Like the Steap proteins, the yeast FRE proteins also contain a predicted flavin-NAD(P)H-binding domain. However, in the latter group, this domain is structurally distinct, and located C-terminal, rather than N-terminal, to the predicted transmembrane spans (Fig. 6a).
Figure 6.
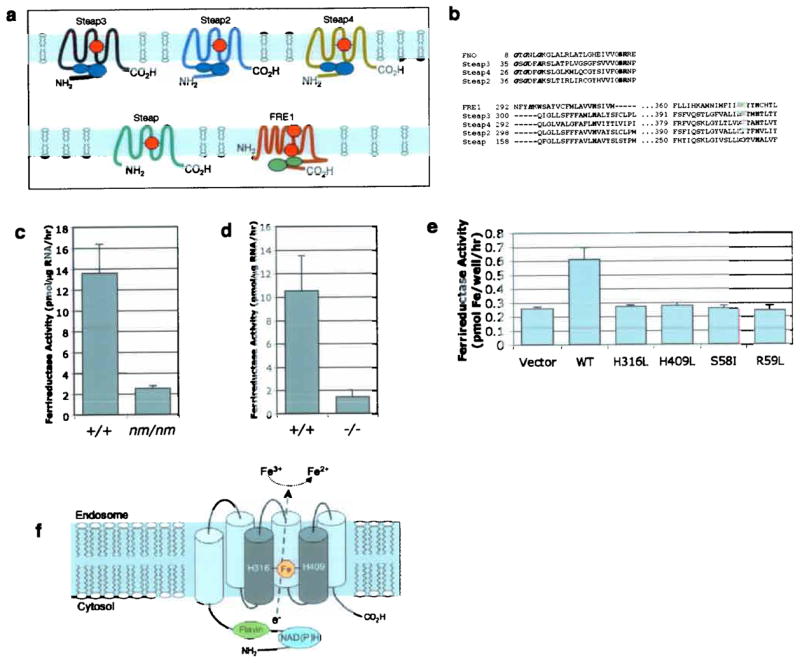
Structure of the Steap family and function of Steap3 (a) Schematic diagram of Steap1-4 and yeast FRE1. Blue ovals in tandem represent the unique flavin- NAD(P)H binding domain. Heme groups are indicated in red. The green in FRE1 represents the FAD:NAD(P)H domain. (b) (top) Multiple sequence alignment of the flavin F420H2: NADP+ Oxidoreductase (FNO) from Archaeoglobus fulgidus and the Mus musculus Steap family members. Steap lacks the FNO-like domain. Conserved residues in the Rossman fold motif (GXGXXA/G) are in bold italics. Serine (S) and arginine (R) residues critical for binding to the phosphate moiety of NADP+ are in bold. (bottom) Predicted transmembrane domains 3 (TM3) and 5 (TM5) of the Steap proteins are aligned with Saccharomyces cerevisiae FRE1. Conserved histidines (H) residues are in bold. The second, non-conserved histidine in each FRE1 TM is in bold italic. Multiple sequence alignment was performed with MultAlign (http://cbrg.inf.ethz.ch/Server/MultAlign.html). (c) Ferrireductase activity in Steap3+/+ (n=10) and Steap3nm1054/nm1054 (n=5) reticulocyte-rich RBCs (d) Ferrireductase activity in Steap3+/+ (n=6) and Steap3−/− (n=5) reticulocyte-rich RBCs. (e) Ferrireductase activity in HEK 293T cells transfected with vector alone (n=12), or wild type (n=12), or mutant Steap3 (n=6 each) expression constructs. Error bars ± 1 S.D. (f) Schematic model of Steap3 mediated reduction of iron. Putative Steap3 membrane topology, FNO motif, and heme coordination sites located in TM3 and TM5.
Steap3 was initially described as a putative tumor suppressor gene capable of inhibiting tumor cells through a caspase-3 dependent pathway11–13,15. It has subsequently been reported to interact with and facilitate secretion of the pro-inflammatory translationally controlled tumor protein (TCTP, also known as histamine-releasing factor [HRF])14. Steap4 has been characterized as a cell surface protein induced by tumor necrosis factor-alpha (TNF-α) in an adipogenic cell line20. STEAP and STEAP2 are highly expressed in the prostate and have been circumstantially implicated in prostate cancer metastasis21,24,25. However, in no case has a specific enzymatic function been attributed to this family of proteins.
The functional defect in Steap3nm1054 and Steap3−/− reticulocytes, as well as the structural homology to oxidoreductases in archaea and the yeast FRE proteins, suggested that Steap3 might function as a ferrireductase. To address this hypothesis, we measured ferrireductase activity in Steap3nm1054 and Steap3−/− reticulocytes5,26. We found that reticulocytes of both homozygous mutant genotypes were similarly deficient in ferrireductase activity compared to controls (Fig. 6c,d). To determine if Steap3 over-expression could stimulate ferrireductase activity in vitro, we transiently transfected HEK 293T cells with Steap3 and found that ferrireductase activity was increased (Fig. 6e). Mutation of either of two residues implicated in NAD(P)H binding (S58I and R59L) completely abrogated this activity (Fig. 6e). A similar loss of function was also seen in Steap3 mutants in which either of the two putative heme-binding histidines was changed to leucine (H316L or H409L) (Fig. 6e). In each case, the mutations had no effect on protein expression or localization (not shown). These results demonstrate the importance of these conserved motifs for reductase activity, and argue against an indirect effect of Steap3 in inducing another ferrireductase protein.
Steap3 has structural features distinct from other known ferrireductases. Within eukaryotes, the F420H2:NADP+ oxidoreductase domain is unique to members of the Steap family (Supplementary Fig. 3 online). Prior studies of the Steap proteins predict a cytoplasmic orientation for the oxidoreductase domain20,21,24. In such a configuration, this domain would have access to the cytoplasmic pool of NAD(P)H as a source of electrons for the reduction of iron (Fig. 6f). In the case of the yeast FRE proteins, the transfer of electrons from NADPH across the membrane is believed to occur sequentially, first through FAD, and then through two heme molecules27. These heme moieties are coordinated by 4 histidine residues within the transmembrane segments22 In Steap3 and its mammalian homologues, only two of these histidine residues are conserved, predicting one intramembrane heme molecule (Fig. 6a,b). Computational modeling of the transmembrane domains of Steap proteins places the single heme more centrally within the membrane (TMpred http://www.ch.embnet.org/software/TMPRED); whereas in the yeast FRE proteins the heme groups are arranged in tandem, at opposite poles of the transmembrane helices (Fig. 6a,b,f)18
Overall, the modeling and in vitro ferrireductase activity of Steap3 would predict that Steap3 binds NAD(P)H and a flavin derivative as well as one heme, and that the reduction of iron is accomplished by sequential transfer of electrons from the NAD(P)H oxido-reductase domain, to an intra-membrane heme cofactor, and ultimately to Fe3+ within the endosome. It is unclear if “free” Fe3+ is the substrate of this reaction, or if reduction facilitates release of iron from Tf9.
Altogether, these data formally define the role of iron reduction in the Tf cycle, and support the hypothesis that Steap3 is an endosomal ferrireductase critical for Tf-Tfr1-mediated iron uptake in erythroid cells. Since Steap3 is found in abundance in other tissues that express high levels of Tfr1, including placenta, the absence of pathologic phenotypes of iron metabolism attributable to these tissues suggests functional overlap in the ferrireduction system. As the other Steap2 and Steap4 are highly homologous to Steap3 and contain oxidoreductase domains, the requirement for an iron reductase in other organs could potentially be met by other family members.
Methods
Genetic and Physical mapping of nm1054
All animal work was reviewed and approved by the Animal Care and Use Committee at Children’s Hospital Boston. A total of 542 affected [CBA/J-nm1054 x CAST/Ei] F2 intercross animals were obtained and genotyped and analyzed as previously described10. Closely linked microsatellite markers D1Mit27 and D1Mit217, located centromerically and telomerically of the deletion, respectively, were used to detect the CBA/J-derived Steap3nm1054 allele thereafter. A redundant overlapping contig of BAC clones was constructed by screening the CITB strain 129S4/SvJae library (Invitrogen) by PCR, and the RPCI-22 strain 129S6/SvEvTac library (http://bacpac.chori.org/home.htm) with gene-specific probes.
Steap3 RNA expression analysis
In situ hybridization was performed on frozen sections of murine tissues using 35S-labelled RNA probes (coding sequence nucleotides 124–519) as previously described28. Real time PCR was performed in triplicate on the Human MTC cDNA Panel I and Human Immune System MTC Panel cDNA sets (BD Biosciences) employing the SYBR Green PCR Master Mix (Stratagene), and the ABI Prism 7700 Sequence Detection System Mx3000P™ Real-Time PCR System according to the manufacturer's instructions. Standard curves were generated using 1, 0.2, 0.1, 0.05, 0.02, and 0.01 ng of total cDNA from human liver. Primers (Supplementary Table 3 online) were designed using the real time PCR primer design software from GeneScript (Piscataway, NJ). Dissociation curve analysis, gel electrophoresis, and sequencing of PCR products were performed to validate and determine the specificity of the assays. The experiment was repeated three times with similar results.
Transient and stable expression of Steap3
The murine Steap3 cDNA was cloned into pCDNA6-myc-HisA (Invitrogen) to create pCDNA6-Steap3-Myc-His. Mutations were introduced by using the QuickChange II site-directed mutagenesis kit (Stratagene). The C-terminally Myc-tagged Steap3 was subcloned from pCDNA6-Steap3-Myc-His into the retroviral vector pMSCVneoEB to generate pMSCV-Steap3-Myc. Tissue culture cells were maintained in DMEM (Gibco-BRL) supplemented with 10% FBS (Invitrogen) and penicillin/streptomycin. Stable expression cell lines were obtained by transfecting HEK 293T or COS-7 cells with linearized pCDNA6-Steap3-Myc-His using Geneporter2 (Gene Therapy Systems), and culturing with blasticidin antibiotic selection. Clones were chosen on the basis of antibiotic resistance, as well as by protein expression determined by western blot and immunofluorescence.
Sub-cellular localization
HEK 293T clones stably expressing low levels of the Steap3-Myc-His protein were plated on poly-D-lysine coated coverslips (Becton-Dickinson) and incubated in DMEM supplemented with 10% FBS and 10μg/ml human holo-Tf (Sigma) for 24 hours prior to immunofluorescence. For Tf labeling, cells were incubated with 30μg/ml Alexafluor 488-labelled human holo-Tf (Molecular Probes) for 30 minutes before washing three times with PBS. Cells were then fixed and treated with a Cy3-labelled mouse anti-Myc 9E10 monoclonal antibody (Sigma) before viewing by confocal microscopy. For Tf receptor co-localization, cells were stained with FITC-conjugated anti-TfR (clone #M-A712, BD Pharmingen). A C-terminally flag tagged DMT1 in pCS2 was transfected into a 293T clone stably expressed myc-tagged Steap3. Cells were stained as previously described29.
Retroviral complementation
Retroviral production, infection, and transplantation were modified from established procedures30. Briefly, retroviral supernatants were harvested from HEK 293T cells 48 hours post-transfection with pMSCVneoEB retrovirus constructs and Ecopac (Cell Genesys, Foster City, CA). C57BL/6J-nm/nm Gpi1b/b [N19] fetal liver hematopoietic cells were obtained at E14.5-16.5. Mature erythrocytes were lysed in 150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA [pH 7.4], and the nucleated fetal liver cells incubated for 24 hours in RPMI supplemented with murine IL-3 (6 U/ml; R&D Systems), recombinant murine stem cell factor (SCF, 5 U/ml; Stem Cell Technologies), recombinant murine IL-6 (10 000 U/ml; R&D Systems), 10% FBS, and 100 U/ml penicillin/streptomycin, and transduced twice at 24-hour intervals with retroviral supernatants. Following retroviral transduction2x106 nucleated fetal liver cells were transplanted by retro-orbital injection into lethally irradiated C57BL/6J-Gpi1a/a mice (Jackson Labs, Bar Harbor, ME). Peripheral blood was collected for phenotypic and chimerism (Gpi1b donor vs. Gpi1a recipient) analysis at 4 weeks post-transplantation as previously described10.
Gene targeting, allelism testing, and phenotyping
BAC transgenics were created by injecting pronuclei of C57BL/6 blastocysts with circular BAC DNA prepared with the NucleoBond BAC Maxi Kit (BD Biosciences). ES cell (J1 line; 129S4/SvJae derivation) gene targeting, culture, and blastocyst injections were performed using standard techniques. Chimeric Steap3+/− males were bred to 129S6/SvEvTac-nm/+ [N5] (Steap3nm1054/+) females, and compound heterozygous (Steap3nm1054/−) and heterozygous (Steap3nm1054/+ and Steap3+/−) F1 animals were intercrossed to produce F2 litters, which were phenotyped as previously described10. Routine genotypes and phenotypes were determined by a PCR assay for the targeted and wild type alleles and by a peripheral blood smear. Statistical analysis was performed using StatView 5.0.1 (SAS Institute Inc, Cary, NC)
Iron uptake and iron reductase experiments
Iron uptake experiments were performed on reticulocyte rich RBCs as previously described10. Reductase assays employed HEK 293T cells in 12 well dishes. Cells were transfected with 1μg of pCDNA6-Steap3-Myc-His or an anti-sense construct using Geneporter2 (Gene Therapy Systems) according to the manufacturer’s instructions. For the assay, transfected cells were incubated in PBS supplemented with 5.6 mM glucose containing 50μM Fe3+- nitrilotriacetic acid (Fe-NTA; 50μM ferric chloride + 100μM nitrilotriacetic acid) and 200μM ferrozine5. After incubation at 37°C for 60 minutes, the Fe2+-ferrozine complex was detected by measuring the absorbance at 562 nm, and converted to Fe2+ generated by using the extinction coefficient, 27.9 mM−1cm−1. Pilot experiments demonstrated time and temperature dependence of the assay. For reticulocyte experiments, a reticulocytosis was induced by bleeding10, and washed reticulocyte-rich RBCs were incubated in Hank’s Balanced Salt Solution (HBSS) containing 50μM Fe3+- nitrilotriacetic acid and 200μM ferrozine for 30 minutes. Iron uptake and ferrireductase activity were normalized to RNA content10
Supplementary Material
Acknowledgments
The authors thank the ongoing support and criticism of Lance Lee, Hiromi Gunshin, Nancy Andrews, Ellis Neufeld, and the members of the Andrews and Neufeld Laboratories at Children’s Hospital Boston. Nicole Stokes and Tiffany Borjeson are thanked for technical support. Supported by the Pew Biomedical Scholars Program (M.D.F), NIH R01 HL074247 (M.D.F.), and NIH R01 DK27726 (J.E.B.). Transgenic core facilities were supported by NIH P30 DK49216.
Footnotes
Competing Interest Statement: The authors declare that they have no competing financial interests.
References
- 1.Ponka P. Tissue-specific regulation of iron metabolism and heme synthesis: distinct control mechanisms in erythroid cells. Blood. 1997;89:1–25. [PubMed] [Google Scholar]
- 2.Fleming MD, et al. Microcytic anemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 3.Fleming MD, et al. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci U S A. 1998;95:1148–53. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunshin H, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 5.McKie AT, et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–1759. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- 6.Gunshin H, et al. in preparation. [Google Scholar]
- 7.Inman RS, Coughlan MM, Wessling-Resnick M. Extracellular ferrireductase activity of K562 cells is coupled to transferrin-independent iron transport. Biochemistry. 1994;33:11850–11857. doi: 10.1021/bi00205a022. [DOI] [PubMed] [Google Scholar]
- 8.Nunez MT, Gaete V, Watkins JA, Glass J. Mobilization of iron from endocytic vesicles. The effects of acidification and reduction. Journal of Biological Chemistry. 1990;265:6688–92. [PubMed] [Google Scholar]
- 9.Dhungana S, et al. Redox properties of human transferrin bound to its receptor. Biochemistry. 2004;43:205–9. doi: 10.1021/bi0353631. [DOI] [PubMed] [Google Scholar]
- 10.Ohgami RS, et al. nm1054, a spontaneous, recessive, hypochromic, microcytic anemia mutation in the mouse. Blood. 2005 doi: 10.1182/blood-2005-01-0379. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porkka KP, Nupponen NN, Tammela TL, Vessella RL, Visakorpi T. Human pHyde is not a classical tumor suppressor gene in prostate cancer. Int J Cancer. 2003;106:729–35. doi: 10.1002/ijc.11278. [DOI] [PubMed] [Google Scholar]
- 12.Steiner MS, Zhang X, Wang Y, Lu Y. Growth inhibition of prostate cancer by an adenovirus expressing a novel tumor suppressor gene, pHyde. Cancer Res. 2000;60:4419–25. [PubMed] [Google Scholar]
- 13.Zhang X, Steiner MS, Rinaldy A, Lu Y. Apoptosis induction in prostate cancer cells by a novel gene product, pHyde, involves caspase-3. Oncogene. 2001;20:5982–90. doi: 10.1038/sj.onc.1204831. [DOI] [PubMed] [Google Scholar]
- 14.Amzallag N, et al. TSAP6 facilitates the secretion of translationally controlled tumor protein/histamine-releasing factor via a nonclassical pathway. J Biol Chem. 2004;279:46104–12. doi: 10.1074/jbc.M404850200. [DOI] [PubMed] [Google Scholar]
- 15.Passer BJ, et al. The p53-inducible TSAP6 gene product regulates apoptosis and the cell cycle and interacts with Nix and the Myt1 kinase. Proc Natl Acad Sci U S A. 2003;100:2284–9. doi: 10.1073/pnas.0530298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labbe R, Vreman H, Stevenson D. Zinc protoporhyrin: A metabolite with a mission. Clin Chem. 1999;45:2060–72. [PubMed] [Google Scholar]
- 17.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–41. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Pulido L, Rojas AM, Valencia A, Martinez AC, Andrade MA. ACRATA: a novel electron transfer domain associated to apoptosis and cancer. BMC Cancer. 2004;4:98. doi: 10.1186/1471-2407-4-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warkentin E, et al. Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound. Embo J. 2001;20:6561–9. doi: 10.1093/emboj/20.23.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moldes M, et al. Tumor necrosis factor-alpha-induced adipose-related protein (TIARP), a cell-surface protein that is highly induced by tumor necrosis factor-alpha and adipose conversion. J Biol Chem. 2001;276:33938–46. doi: 10.1074/jbc.M105726200. [DOI] [PubMed] [Google Scholar]
- 21.Porkka KP, Helenius MA, Visakorpi T. Cloning and characterization of a novel six-transmembrane protein STEAP2, expressed in normal and malignant prostate. Lab Invest. 2002;82:1573–82. doi: 10.1097/01.lab.0000038554.26102.c6. [DOI] [PubMed] [Google Scholar]
- 22.Shatwell KP, Dancis A, Cross AR, Klausner RD, Segal AW. The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J Biol Chem. 1996;271:14240–4. doi: 10.1074/jbc.271.24.14240. [DOI] [PubMed] [Google Scholar]
- 23.Finegold AA, Shatwell KP, Segal AW, Klausner RD, Dancis A. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J Biol Chem. 1996;271:31021–4. doi: 10.1074/jbc.271.49.31021. [DOI] [PubMed] [Google Scholar]
- 24.Hubert RS, et al. STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci U S A. 1999;96:14523–8. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang D, Holt GE, Velders MP, Kwon ED, Kast WM. Murine six-transmembrane epithelial antigen of the prostate, prostate stem cell antigen, and prostate-specific membrane antigen: prostate-specific cell-surface antigens highly expressed in prostate cancer of transgenic adenocarcinoma mouse prostate mice. Cancer Res. 2001;61:5857–60. [PubMed] [Google Scholar]
- 26.Goldman GL, Thornton JI. A new trace ferrous metal detection reagent. J Forensic Sci. 1976;21:625–8. [PubMed] [Google Scholar]
- 27.Lesuisse E, Casteras-Simon M, Labbe P. Evidence for the Saccharomyces cerevisiae ferrireductase system being a multicomponent electron transport chain. J Biol Chem. 1996;271:13578–83. doi: 10.1074/jbc.271.23.13578. [DOI] [PubMed] [Google Scholar]
- 28.Ross JS, et al. Correlation of primary tumor prostate-specific membrane antigen expression with disease recurrence in prostate cancer. Clin Cancer Res. 2003;9:6357–62. [PubMed] [Google Scholar]
- 29.Su MA, Trenor CC, Fleming JC, Fleming MD, Andrews NC. The G185R mutation disrupts function of iron transporter Nramp2. Blood. 1998;92:2157–63. [PubMed] [Google Scholar]
- 30.Schwaller J, et al. Transformation of hematopoietic cell lines to growth-factor independence and induction of a fatal myelo- and lymphoproliferative disease in mice by retrovirally transduced TEL/JAK2 fusion genes. Embo J. 1998;17:5321–33. doi: 10.1093/emboj/17.18.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


