Abstract
We investigated the nuclear higher order compartmentalization of chromatin according to its replication timing (Ferreira et al. 1997) and the relations of this compartmentalization to chromosome structure and the spatial organization of transcription. Our aim was to provide a comprehensive and integrated view on the relations between chromosome structure and functional nuclear architecture. Using different mammalian cell types, we show that distinct higher order compartments whose DNA displays a specific replication timing are stably maintained during all interphase stages. The organizational principle is clonally inherited. We directly demonstrate the presence of polar chromosome territories that align to build up higher order compartments, as previously suggested (Ferreira et al. 1997). Polar chromosome territories display a specific orientation of early and late replicating subregions that correspond to R- or G/C-bands of mitotic chromosomes. Higher order compartments containing G/C-bands replicating during the second half of the S phase display no transcriptional activity detectable by BrUTP pulse labeling and show no evidence of transcriptional competence. Transcriptionally competent and active chromatin is confined to a coherent compartment within the nuclear interior that comprises early replicating R-band sequences. As a whole, the data provide an integrated view on chromosome structure, nuclear higher order compartmentalization, and their relation to the spatial organization of functional nuclear processes.
Keywords: nuclear architecture, genome organization, chromosome structure, nuclear compartments, polar chromosome territories
Since it became evident that cell nuclei are compartmentalized organelles (for review see Spector 1993; Cremer et al. 1995; van Driel et al. 1995; Strouboulis and Wolffe 1996; Singer and Green 1997; Lamond and Earnshaw 1998), one of the major goals in cell biology has been to understand their functional organization. In particular, the functional organization of genomes within cell nuclei is a topic of long lasting and controversial discussions (Blobel 1985; Hutchinson and Weintraub 1985; Hochstrasser and Sedat 1987; Cremer et al. 1988, Cremer et al. 1995; Manuelidis 1990; Belmont and Bruce 1994; De Boni 1994; Berezney et al. 1995; Kurz et al. 1996; Marshall et al. 1997a; Bridger and Bickmore 1998). Although it has become obvious that genomes are compartmentalized at the level of whole chromosome territories in animal as well as in plant nuclei (Manuelidis 1985; Schardin et al. 1985; Cremer et al. 1988; Lichter et al. 1988; Leitch et al. 1990), it was difficult to understand the internal structure of chromosome territories and the relation of their organization to presumptive higher order functional compartments within nuclei (for review see Cremer et al. 1995; van Driel et al. 1995; Bridger and Bickmore 1998).
Studies on mitotic chromosomes have indicated a functionally significant compartmentalization of mammalian genomes, strongly related to the well known banding patterns and their specific isochore compositions. A correlation of isochores (for review see Bernardi 1995) from GC poor and GC rich families with G- and R-bands (Bernardi et al. 1985, 1989) was demonstrated (Saccone et al. 1993; Federico et al. 1998). Since GC poor and GC rich isochores have low and high gene concentrations, respectively (Cuny et al. 1981; Saccone et al. 1996; Federico et al. 1998), these results also gave an estimate of gene distribution on chromosomal bands.
Two further pieces of evidence suggest that the classical-banded structure observed only on mitotic chromosomes is relevant for genome organization with regard to important nuclear functions like transcription and replication. First, constitutively expressed, housekeeping genes reside almost exclusively within the R-bands, whereas G-bands harbor predominantly tissue-specific genes (Craig and Bickmore 1993, Craig and Bickmore 1994). Second, R-band DNA replicates first during S phase, whereas G-band DNA replicates thereafter, during the second half of S phase (Kim et al. 1975; Dutrillaux et al. 1976; Camargo and Cervenka 1982). Recent studies indicate that the structures of mitotic chromosomes and nuclear chromosome territories are closely related and that the different bands of mitotic chromosomes are present as distinct domains regarded as subchromosomal foci (SF)1 within chromosome territories (Zink et al. 1998, Zink et al. 1999).
Moreover, there are several lines of evidence indicating, on the DNA level, a close relationship between SF and replication foci (RF) observed during S phase (Meng and Berezney, 1991; Sparvoli et al. 1994; Berezney et al. 1995; Jackson and Pombo 1998; Ma et al. 1998; Zink et al. 1998, Zink et al. 1999). RF contain the actively replicated DNA, the nascent DNA, and all factors necessary for replication (Leonhardt and Cardoso 1995). RF are organized into higher order patterns within the nucleus, and studies on fixed cells indicate that there are characteristic patterns for different stages of S phase (Nakayasu and Berezney 1989; O'Keefe et al. 1992; Ferreira et al. 1997). Recent data indicate that higher order nuclear compartments, comprising DNA with a specific replication timing revealed by specific nuclear patterns of foci, are not established during S phase but directly after mitosis at late telophase/early G1 (Ferreira et al. 1997). However, the functional significance of these higher order compartments and their stability during interphase was not clear.
As a whole, the data described above suggest a close relationship between chromosome structure during mitosis and interphase, functional nuclear processes like replication and transcription, and nuclear higher order compartmentalization. However, because of the lack of direct evidence for these relations it was difficult to obtain an integrated view of mammalian genome functional architecture. Therefore, it was important to study the different parameters all in relation to one another. Our aim was to enable for an integration by relating chromosome structure during mitosis and interphase to nuclear higher order compartments characterized by a specific replication timing and to the spatial organization of transcription. We confirm that mammalian genomes are organized into higher order nuclear compartments that harbor DNA sequences with a specific replication timing (Ferreira et al. 1997). We demonstrate that higher order compartments established immediately after mitosis are stably maintained during all interphase stages. The organizational principle is clonally inherited. We provide direct evidence that higher order compartments are built up by the alignment of polar chromosome territories, as suggested by Ferreira et al. 1997. Polar chromosome territories display clusters of early or late replicating DNA that correspond to R- and G/C-bands of mitotic chromosomes, respectively, at distinct subterritorial positions. The R-band sequences of distinct chromosomes cluster within the nuclear interior to give rise to an early replicating compartment that is transcriptionally competent and active. The G/C-band sequences organize transcriptionally incompetent and inactive late replicating compartments in the perinuclear and perinucleolar regions.
Materials and Methods
Cell Culture
Cells were grown at 37°C in an atmosphere of 5% CO2 in RPMI supplemented with 10% FCS (HeLa S6 cells [provided by W.W. Franke, DKFZ, Heidelberg]), female human diploid fibroblasts (Hv provided by Professor J. Murken, LMU, Munich), SH-EP-N14 human neuroblastoma cells (derivative of SH-EP-cells [Ross et al. 1983], stably transfected with an N-Myc expression vector [Wenzel et al. 1991]) and CHO cells (provided by S. Müller, LMU, Munich). Primary human lymphocytes and C2C12 mouse myoblasts (provided by M.C. Cardoso, MDC, Berlin) were cultured in the presence of 20% FCS. The media were supplemented with antibiotics (100 μg/ml penicillin and 100 μg/ml streptomycin). 4 μg/ml phytohemagglutinin were added to cultures of primary lymphocytes. For synchronization in early S phase, 200 μM mimosin was added to the culture medium for 14–16 h. The block was released by adding fresh medium after washing the cells with PBS. For microinjection and microscopy of fixed cells, cells were grown on coverslips and fixed after replication labeling/nascent RNA labeling in PBS/3.7% formaldehyde for 10 min. Fixed cells were stored in PBS at 4°C and always kept wet during all of the following immunostaining/in situ hybridization procedures. For following the clonal inheritance of replication labeling patterns in living cells, the cells were cultured, microinjected, and imaged in cell culture dishes with a gridded coverslip (cellocate, Eppendorf-Netheler-Hinz GmbH) inserted into the bottom.
Replication Labeling
Replication labeling was performed with exponentially growing cultures or cultures synchronized in early S phase. Replication labeling involving a single BrdU (bromodeoxyuridine) or Cy3-dUTP pulse (Cy3-dUTP was microinjected) or iododeoxyuridine (IdU)/chlorodeoxyuridine (CldU) double pulse labeling was performed as described in (Zink et al. 1998) according to the time schedules described in the results. FITC-dUTP (Boehringer) was microinjected at a concentration of 100 μM diluted in CMF-PBS (PBS without Ca++ and Mg++). If synchronized cultures were used, the block was released either after microinjection or before BrdU or IdU was supplemented to the medium to obtain early S phase patterns. To obtain later S phase patterns, the cells were replication-labeled 2–9 h after release. Cells were fixed at the time points indicated in the results either during or after S phase. If necessary, detection of incorporated nucleotides was performed as described in (Zink et al. 1998, Zink et al. 1999) (in the present study BrdU was only detected with a mouse anti–BrdU antibody and an anti-mouse TRITC-conjugated secondary antibody, see BrUTP detection). No detection procedures were necessary for visualizing incorporated fluorescent nucleotides (Cy3-dUTP, FITC-dUTP).
Replication Labeling in Combination with Nascent RNA Labeling
Synchronized or unsynchronized cells from cultures of HeLa S6 or CHO cells were replication-labeled by microinjection of FITC-dUTP and grown after replication labeling for 14–19 h. Subsequently, replication-labeled cells were microinjected with 5-bromouridine-5′-triphosphate (BrUTP, Sigma Chemical Co., 50 mg/ml in CMF-PBS). After 10 min of microinjection, cells were fixed. For immunodetection of BrUTP, preparations were blocked for 30 min in PBS, 0.3% Triton X-100, 0.2% Tween 20, and 3% BSA, and subsequently incubated with a monoclonal anti–BrdU antibody (Becton and Dickinson, recognizes also BrUTP) diluted in blocking solution for 1 h. After washing the preparations three times for 5 min with PBS, 0.2% Triton cells were incubated with the secondary antibody (TRITC conjugated goat anti–mouse; Dianova) diluted in the blocking solution. After washing the cells three times for 5 min in PBS, preparations were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml in PBS) and mounted (Vectashield) for microscopy.
Replication Labeling in Combination with Immunostaining Against Hyperacetylated Histone H4
Cells were replication-labeled either with Cy3-dUTP or BrdU and fixed. Fixed preparations were permeabilized and blocked in blocking solution (PBS, 0.2% Triton X-100, 0.2% Tween 20, 5% BSA) at room temperature for 1 h. Subsequently, cells were incubated with rabbit antiserum R 232/8 diluted 1:500 in blocking solution for 1 h at room temperature. R 232/8 is a high titer rabbit antiserum that is specific for histone H4 acetylated at lysine 8. It recognizes more highly acetylated H4 isoforms (mainly di- and triacetylated isoforms). Cells were washed three times for 10 min with PBS and 0.2% Triton X-100. Afterwards, cells were incubated with an FITC-conjugated goat anti–rabbit antibody (Dianova) diluted in blocking solution. Cells were washed three times for 10 min with PBS and mounted for microscopy if they were replication-labeled with Cy3-dUTP. In the case when cells were replication-labeled with BrdU, they were postfixed with PBS, 3.7% formaldehyde for 10 min. After fixation, DNA was denatured for BrdU detection by incubating the preparations for 10 min in 2 M HCl (for BrdU detection procedure see above).
Preparation of the H3 Isochore Probe for In Situ Hybridization
DNA from an H3 isochore fraction of human DNA (equivalent to one previously described, Saccone et al. 1996) was amplified and labeled by DOP-PCR (Telenius et al. 1992) with biotin-16-dUTP (Boehringer). Fragment length was checked by gel electrophoresis after DNase I digestion (3 μg/ml for ∼30 min at 15°C). 200 (for metaphase spreads) or 400 ng (for interphase nuclei) of labeled DNA was precipitated with 30 μl CotI-DNA (1 mg/ml; GIBCO BRL) and 5 μl of salmon testis DNA (11 mg/ml; GIBCO BRL). The dry pellet was dissolved in 10-μl hybridization solution (50% formamide, 10% dextran sulfate, 1× SSC). Before hybridization, the probe DNA was denatured in hybridization solution for 6 min at 75°C and preannealed for 20 min at 37°C.
In Situ Hybridization and Probe Detection
Metaphase spreads of SH-EP-N14 cells and primary lymphocytes were prepared according to standard protocols (Macgregor and Varley 1988). After denaturation (2 min at 72°C in 70% formamide, 0.6× SSC, pH 7), the preparations were dehydrated in an ethanol series and air dried. Denatured probe in hybridization solution was applied and hybridized overnight at 37°C under a sealed coverslip. Fixed interphase nuclei were pretreated for hybridization and hybridized as described in Zink et al. 1998. Detection of hybridized probe DNA followed the same procedure for metaphase spreads and interphase nuclei. Preparations were washed for 5 min in 2× SSC at 37°C and three times for 5 min in 0.1× SSC at 60°C and blocked for 1 h at 37°C in 4× SSC, 0.2% Tween 20 (SSCT) with 3% BSA. Cells were incubated with fluorescein avidin DCS (1:200 in SSCT, 1% BSA; Vector Labs, Inc.) for 45 min at 37°C. Cells were washed three times for 10 min with SSCT and incubated for 45 min with a biotinylated anti–avidin antibody (1:200 in SSCT, 1% BSA; Vector Labs, Inc.). After washing the preparations three times for 10 min in SSCT they were incubated again with fluorescein avidin DCS as described above. Cells were finally washed three times for 5 min in SSCT, counterstained with DAPI, and mounted for microscopy (see above). Cells prepared for in situ hybridization were only replication-labeled with Cy3-dUTP.
Imaging
Confocal imaging of nuclei was performed as described in Eils et al. 1996. Epifluorescence microscopy was performed with an Axiovert microscope 135 TV (Zeiss) equipped with an Attoarc device (Zeiss) to regulate light intensity during living cell imaging and a CCD camera (MicroMAX; Princeton Instruments). For imaging, the Metamorph software (version 3.0; Universal Imaging Corp.) was used. Images were arranged using Adobe Photoshop (version 4.0).
Results
Higher Order Compartments Comprising DNA with a Defined Replication Timing Are Present during All Interphase Stages
Previous studies suggested a nuclear higher order compartmentalization of mammalian genomes according to the replication timing of DNA sequences (Ferreira et al. 1997). The nuclear compartments appear to be established at late telophase/early G1 (Ferreira et al. 1997). However, the stability of nuclear compartments established after mitosis during subsequent interphase stages and the stability of their inheritance was not clear. We were interested in the question of whether compartments comprising DNA sequences with a similar replication timing are indeed reproducibly established after mitosis and stably maintained during all subsequent interphase stages. In this case, a specific S phase pattern produced by DNA pulse labeling during replication should be observed at all other interphase stages and in all daughter cell nuclei.
To prove this prediction, we pulse-labeled DNA of different cell lines and primary cells derived from man, mouse, and hamster during the S phase. Studies were performed with the following cell types: primary human diploid fibroblasts (HDFs), HeLa S6 cells, human neuroblastoma cells (SH-EP N14), CHO cells, and C2C12 mouse myoblasts. Pulse labeling was performed with BrdU or Cy3-dUTP. Cells were fixed 30 min after supplementing with the modified nucleotides to obtain typical S phase patterns (Fig. 1, left, a, c, e, g, and i).
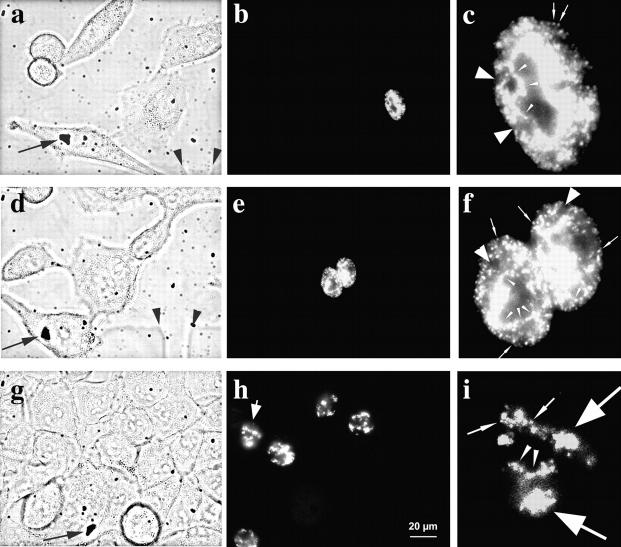
Figure 1.
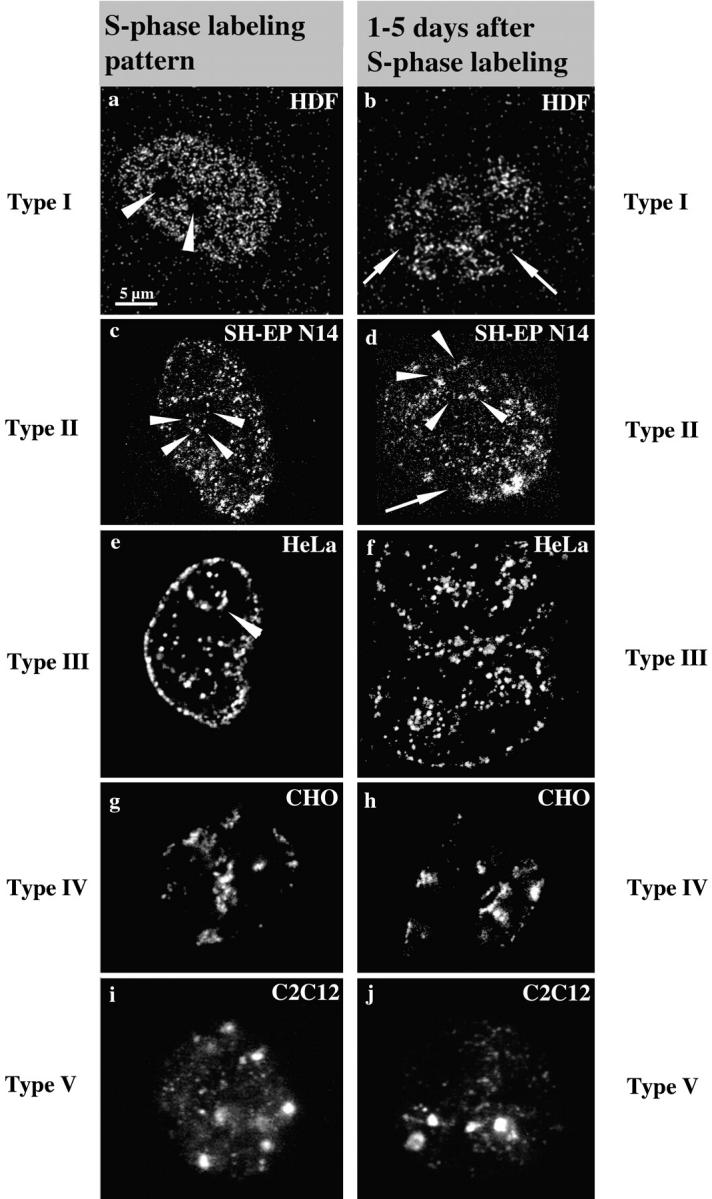
S phase replication labeling patterns are preserved. Cells ([a and b] Hv, human diploid fibroblasts; [c and d] SH-EP N14 human neuroblastoma cells; [e and f] HeLa cells; [g and h] CHO cells; and [i and j] C2C12 mouse myoblasts) were replication-labeled with BrdU (a–d, i, and j; 30-min pulses) or Cy3-dUTP (e–h). Cells were fixed immediately after BrdU labeling or 30 min after microinjection of Cy3-dUTP to obtain labeled S phase cells (left, a, c, e, g, and i). S phase cells display the typical replication labeling patterns (indicated on the left, for classification see text): (a) type I, (c) type II, (e) type III, (g) type IV, and (i) type V. Similarly labeled cells were grown for 1–5 d after labeling and fixed after this growth period (right, b, d, f, h, and j). The presence of similar patterns (types indicated on the right) several days after S phase labeling suggests that DNA replicating with a particular timing during S phase always occupies similar nuclear positions. Arrowheads in a mark the nucleoli that are always unlabeled. Arrowheads in c–e mark perinucleolar label. Arrows in b and d indicate unlabeled chromosome territories. The presence of unlabeled territories demonstrates that cells do not display similar patterns because they stopped cycling, but rather that they went through at least two mitoses after labeling (compare Fig. 3). Note also the two very close nuclei in f displaying similar type III patterns, suggesting that similar patterns were conserved in sister nuclei after cell division (compare Fig. 4, a–c). All pictures display single light optical sections through midnuclear planes except i and j, which show epifluorescence images. Bar in a is similar for all images.
Regarding S phase patterns, we mainly followed the classification of O'Keefe et al. 1992 and defined five different types of pattern reflecting an ordered time sequence during S phase progression (type I beginning and type V end of S phase): type I (Fig. 1 a) displays hundreds of small foci (∼300 nm in diameter) distributed throughout the DNA within the nuclear interior. DNA sequences located at the nuclear and nucleolar peripheries and minor accumulations of late replicating chromatin within the nuclear interior (see type IV and V patterns) are not labeled. Nucleoli are excluded in all types of patterns. The nuclear space occupied by the type I pattern will be regarded as the interior compartment in the following text.
The type II pattern (Fig. 1 c) shows some labeling of DNA located at nuclear and nucleolar peripheries. The interior compartment is still partially labeled, but fewer foci are observed here compared with the type I pattern and unlabeled regions appear within the interior compartment. The type III pattern (Fig. 1 e) displays heavy labeling of nuclear and nucleolar peripheries (excluded from the type I pattern), whereas the interior compartment is almost devoid of label. The compartments labeled by the type III pattern will be regarded as the peripheral compartments.
Type IV and V (Fig. 1g and Fig. i) patterns are characterized by a few large (∼800 nm in diameter) foci that are located within the nuclear interior as well as in the peripheral compartments. Type V displays less labeling of the peripheral compartments than type IV. The totality of late replicating chromatin accumulations within a nucleus labeled by type IV and V patterns will be regarded as the late replicating compartments in the following text. All cell lines examined displayed type I–V patterns.
These results confirmed for all cell types used the finding that DNA sequences with a defined replication timing occupy during S phase specific nuclear areas (Nakayasu and Berezney 1989; O'Keefe et al. 1992). This way, higher order nuclear compartments are established, comprising DNA with a similar replication timing (e.g., the interior compartment comprises the early replicating DNA sequences). To investigate the stability of these compartments during cell growth and distinct cell cycle stages, we pulse-labeled cells during the S phase as described above. In contrast to the previous experiment, cells were not fixed immediately but after a growth period of 1–5 d. The labeling patterns after this growth period were compared with typical S phase patterns (Fig. 1).
The patterns we observed 1–5 d after labeling (Fig. 1, right, b, d, f, h, and j) were similar to typical S phase patterns (Fig. 1). The distribution of numbers and sizes of foci was the same as that seen in cells fixed during S phase. 200 cells were examined for each cell type and no labeling patterns were observed that could not be classified according to the five types of patterns outlined above. Therefore, we extended the classification of the five types of patterns also to non-S phase cells.
The only difference to typical S phase patterns observed in cells grown for 1–5 d was the appearance of unlabeled nuclear areas corresponding to unlabeled chromosome territories (Fig. 1b and Fig. d). The appearance of unlabeled chromosome territories is due to the random segregation of labeled and unlabeled chromatids beginning at the second mitosis after replication labeling (Taylor 1984; Zink et al. 1998) (see also Fig. 3). This phenomenon confirmed that the cells went through at least two mitoses after labeling, and excluded the possibility that cells maintained the patterns because they stopped cycling. As the patterns were often similar in neighboring cells (Fig. 1 f) the data suggested that nuclear compartments comprising DNA with a similar replication timing were clonally inherited.
Figure 3.
Clonal inheritance of replication labeling patterns. HeLa cells were grown on a gridded cellocate coverslip and the same area (bar in h) was imaged on three consecutive days (a–c, day 1; d–f, day 2; g–i, day 3; a, d, and g, phase-contrast; b, e, and h, Cy3-detection; and c, f, and i, enlarged Cy3-labeled nuclei). Similarity of the imaged areas is indicated by the grid (arrowheads in a and d) and a piece of debris attached to one cell (arrows in a, d, and g). 8 h after microinjection of Cy3-dUTP (a–c), one cell within the imaged area is labeled. An enlargement of the cell shown in b is depicted in c. The cell displays a type II pattern: the perinucleolar region is labeled (small arrowheads) while small foci (arrows) are distributed over broader nucleoplasmic areas although some areas are excluded (large arrowheads). Note that small foci in areas where the nuclei are particularly thick cannot be resolved by epifluorescence microscopy and appear as uniformly brightly stained areas (confocal imaging was not compatible with this type of living cell study). Overnight the cell divided and on the next day (d–f) two daughter cells display a type II pattern. An enlargement of the cells depicted in e is shown in f (small arrowheads, perinucleolar label; large arrowheads, unlabeled regions; and arrows, small nucleoplasmic foci). Each of the daughter cells divided to give rise to four granddaughter cells on the next day (g–i). At the second mitosis after initial labeling, labeled and unlabeled chromatids segregate (Taylor 1984) giving rise to nuclei containing approximately equal amounts of labeled and unlabeled chromosome territories visible as stained or unstained patches within the nuclei depicted in h. Although the pattern appears different because of the presence of unlabeled territories the enlargement (i, nucleus indicated by an arrow in h) still shows a typical type II pattern with foci enriched along the boundary of a nucleolus (arrowheads) as well as distributed over broader nucleoplasmic areas. These foci belong to particular chromosome territories within the nucleoplasm (large arrows). Epifluorescence microscopy resolves the focal substructure of territories (Zink et al. 1998) only where territories are relatively flat (small arrows).
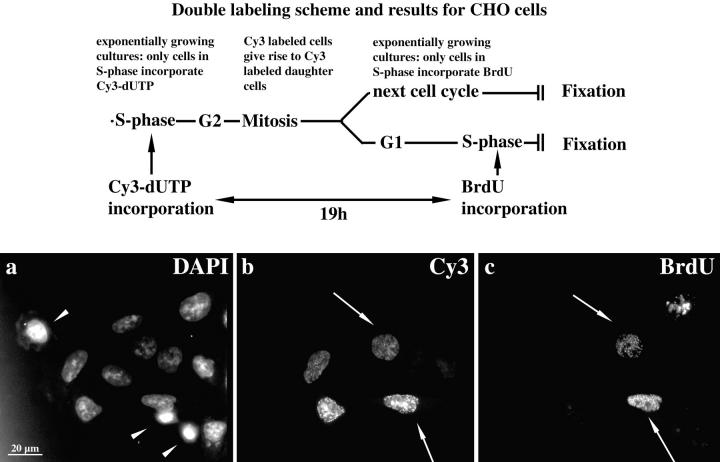
To exclude the possibility that cells displayed labeling patterns reminiscent of S phase patterns because of the fact that cells entered S phase again, exponentially growing CHO cells were pulse-labeled with Cy3-dUTP to obtain typical replication labeling patterns. Cy3-labeled cells were grown for 19 h, pulse-labeled with BrdU for 30 min, and fixed immediately (see scheme depicted in Fig. 2). BrdU labeling revealed whether cells displaying typical type I–V Cy3-patterns had entered S phase again. As the cultures grew exponentially, some daughter cells of Cy3-dUTP replication-labeled cells were in S phase at the time point of fixation (Fig. 2 c), whereas others were not. Nevertheless, all cells displayed the typical type I–V Cy3 labeling patterns (60 cells examined for each cell type), indicating that the corresponding nuclear compartmentalization is present during all interphase stages. Similar results were obtained with HeLa S6 cells.
Figure 2.
Genome compartmentalization is similar during all interphase stages. The applied double labeling procedure is schematically drawn at the top. Typical replication labeling patterns were obtained by incorporation of Cy3-dUTP into the DNA of the S phase cells of exponentially growing CHO cultures. 19 h after Cy3-dUTP microinjection, cultures were replication-labeled with BrdU and fixed after 30 min. Cells in S phase at the time point of fixation (BrdU-labeled) can be distinguished from G1 and G2 cells (not BrdU-labeled). The DAPI staining is shown in a (a–c, same field of cells is imaged). Mitotic stages of exponentially growing cultures are indicated by arrowheads. Cy3-labeled cells are depicted in b. Note the two pairs of cells with similar labeling intensities and labeling patterns (upper pair, type I; lower pair, type III; blurred label is due to epifluorescent imaging). Cells of one pair are likely sister cells (compare Fig. 4). The right cells of each pair were in S phase at the time point of fixation (arrows) as indicated by the BrdU label depicted in c. Cy3 labeling patterns are similar in S phase and non-S phase cells.
Nuclear Genome Compartmentalization Is Stably Inherited through Mitoses
To confirm clonal inheritance of nuclear genome compartmentalization, single cells from HeLa S6 cultures exponentially growing on gridded coverslips were replication-labeled with Cy3-dUTP. Cells were not fixed but imaged each day to follow the inheritance of the initial labeling patterns in developing clones. Unfortunately, the motility of the cells usually did not allow the unequivocal identification of cells belonging to one clone (although usually neighboring cells and fields of cells were observed displaying similar patterns). However, this was possible in the case depicted in Fig. 3 where, indeed, all daughter and granddaughter cells displayed the initial type II labeling pattern.
To further confirm the inheritance of patterns HeLa S6 cells were synchronized with mimosin in early S phase. Immediately after release of the mimosin, block cells were pulse-labeled for 0.5 h with IdU, chased for 9.5 h, and pulse-labeled for 0.5 h with CldU (Fig. 4, labeling scheme). We previously established (Fauth 1998) that 93% of cells (n = 1,000) displayed a type I pattern if they were labeled within the first hour after release. 50% of cells (n = 1,000) displayed a type III pattern and 18% a type IV or V pattern if they were labeled 10 h later after release. Therefore, most cells after IdU/CldU double labeling according to the scheme outlined above, display a type I pattern (labeled with IdU) as well as type III–V pattern (labeled with CldU).
Figure 4.
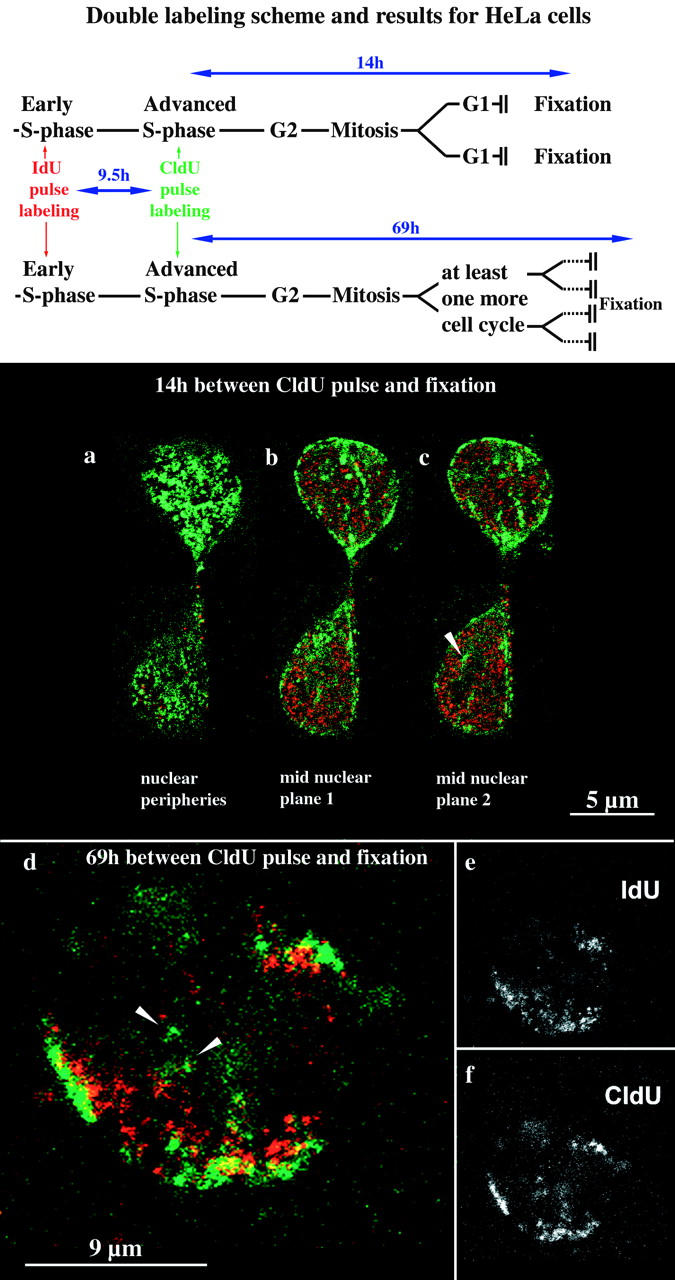
Establishment of higher order nuclear compartments after mitosis and the underlying chromosomal structure. HeLa S6 cells synchronized in early S phase were labeled for 30 min with IdU and 9.5 h later for 30 min with CldU (see labeling scheme at the top). Therefore, initially labeled cells simultaneously displayed a type I pattern labeled by IdU and a pattern typical for the second half of S phase labeled by CldU. Daughter cells of the initially labeled cells were fixed 14 (a–c) or 69 h (d–f), respectively, after the CldU pulse and analyzed by confocal microscopy. Identical nuclear planes were imaged regarding TRITC (IdU detection, depicted in red) and FITC (CldU detection, depicted in green) fluorescence. The corresponding merged TRITC and FITC signals (colocalizing signals appear yellow) are shown in a–d. e and f display only the TRITC (e) or FITC (f) signals of the merged image in d (midnuclear plane). The two cells depicted in a–c display the typical morphology of early G1 cells shortly after mitosis. Both G1 cells display an IdU (red) type I pattern and a CldU (green) type III pattern present in their mother cell. Perinucleolar labeling is indicated by an arrowhead in c. After 69 h (d–f), initially labeled cells went through at least two mitoses as indicated by the presence of single-labeled chromosome territories (double-labeled patches within the nucleus depicted in d). Double-labeled chromosome territories reveal that the nuclear type I (IdU, red) and type III patterns are due to a reproducibly distinct distribution of IdU- and CldU-labeled DNA within single chromosome territories. CldU-labeled DNA is concentrated at subchromosomal positions near the nuclear and nucleolar peripheries, whereas IdU-labeled DNA is located at subchromosomal positions between these compartments.
Cells were fixed 14 or 69 h after the CldU pulse, respectively (Fig. 4, labeling scheme). Many cells fixed 14 h after CldU labeling went through mitosis. After cell division, daughter cells (39 cells examined) displayed the same nuclear compartmentalization as their mother cells after S phase labeling as revealed by the double labeling patterns. For example, cells with the typical morphology of early G1 cells displayed a type I IdU pattern in combination with a CldU type III pattern (Fig. 4, a–c). These data show that nuclear compartments comprising DNA sequences with a similar replication timing are immediately established after mitosis according to the pattern present in the previous interphase.
Polar Chromosome Territories Build Up Higher Order Nuclear Compartments
Similar compartmentalization patterns (IdU type I patterns with CldU type III–V patterns) were also present after at least two mitoses (indicated by the presence of unlabeled chromosome territories) 69 h after initial IdU/CldU labeling. Fig. 4d–f, shows a typical example of the 46 nuclei examined. As single replication-labeled chromosome territories can be observed the data also indicate how single chromosome territories, composed of DNA replicating at distinct time points during S phase, contribute to higher order nuclear compartments. Single territories display a polar organization with DNA replicating early during S phase (IdU-labeled) clustered at subterritorial positions located within the nuclear interior and DNA replicating at later S phase stages (CldU-labeled) clustered at subterritorial positions located at nuclear or nucleolar peripheries. Alignment of polar territories gives rise to higher order nuclear compartments.
Functional Nuclear Compartmentalization of Mammalian Genomes Is Related to the Organization Of Mitotic Chromosomes
R-bands of mitotic chromosomes comprise DNA replicating early during S phase and G- or C-bands harbor DNA replicating at later S phase stages (Kim et al. 1975; Dutrillaux et al. 1976; Camargo and Cervenka 1982). Therefore, one would expect that DNA located within these distinct bands of mitotic chromosomes corresponds to the observed early and late replicating DNA at different subterritorial positions and contributes distinctly to the different higher order compartments. It has been previously shown that clustering of R- and G-band sequences at distinct subchromosomal positions leads to the formation of polar interphase chromosome territories (Zink et al. 1999).
To confirm the contribution of DNA belonging to specific chromosomal bands to distinct higher order nuclear compartments, we performed in situ hybridization in combination with Cy3-dUTP replication labeling. As a probe for in situ hybridization we used the H3 isochore fraction of human DNA (Saccone et al. 1996). The H3 fraction usually hybridizes with a subset of R-bands on human mitotic chromosomes (Saccone et al. 1996). However, when we tested the specificity of hybridization on metaphase spreads of human chromosomes prepared from cultured primary lymphocytes (n = 20; Fig. 5, a and b) as well as SH-EP N14 human neuroblastoma cells (n = 20; data not shown) we obtained almost a complete R-banding pattern although different R-bands were stained with variable intensity (Fig. 5 b). Even though the probe hybridized specifically to the whole set of R-bands, which was contrary to previous results (see Discussion), the result was highly reproducible under the conditions we used.
Figure 5.
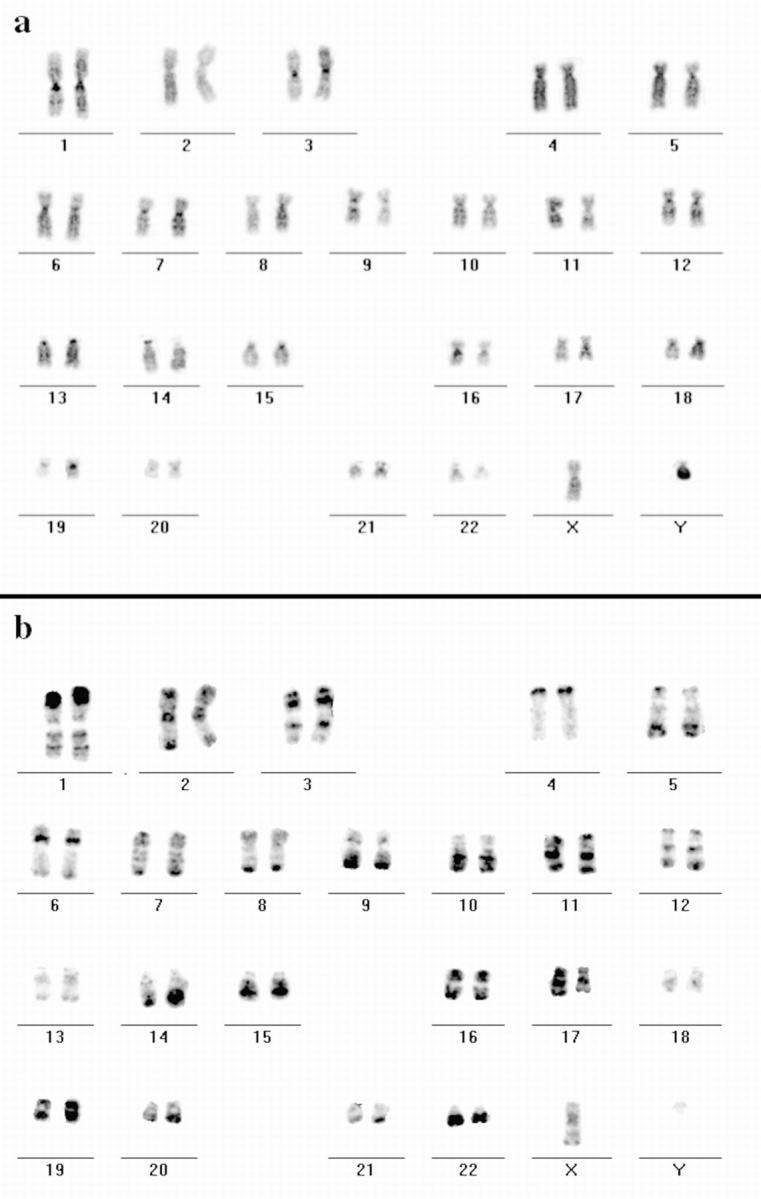
Hybridization of DNA from the H3 isochore fraction to human metaphase spreads. FISH with DNA from the H3 isochore fraction as a probe (FITC-detected) was performed on metaphase spreads from male human lymphocytes. Chromosomes were DAPI-banded and arranged into standard karyograms. The inverted DAPI image (a) displays G- and C-bands more darkly stained compared with R-bands. Most of the R-bands hybridized specifically to the DNA probe as the inverted image of the hybridization signals shows (b, FITC fluorescence appears dark) that displays a typical R banding pattern. Note the different signal intensities of distinct R-bands (e.g., on the distal p-arm of chromosome 1 and the q-arm of chromosome 13).
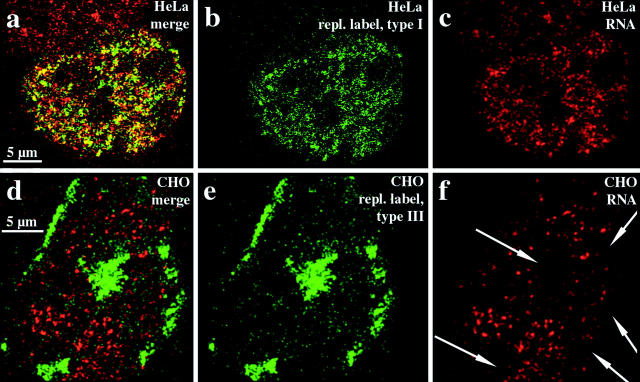
As the probe under the conditions we used was an excellent and specific marker for R-band DNA, we hybridized it to replication-labeled nuclei of HeLa S6 and SH-EP N14 cells (in this case it was not possible to include hamster or mouse cells as the probe is specific for human DNA). The hybridization signal was spread all over the interior compartment (Fig. 6, a–d), but was excluded from the peripheral compartments (Fig. 6 e) and the late replicating compartment (Fig. 6 f). The data confirm that R-band DNA builds up the interior compartment, and reveal a clear correlation between the organization of mammalian genomes during interphase and the banded organization of mitotic chromosomes (see Fig. 10).
Figure 6.
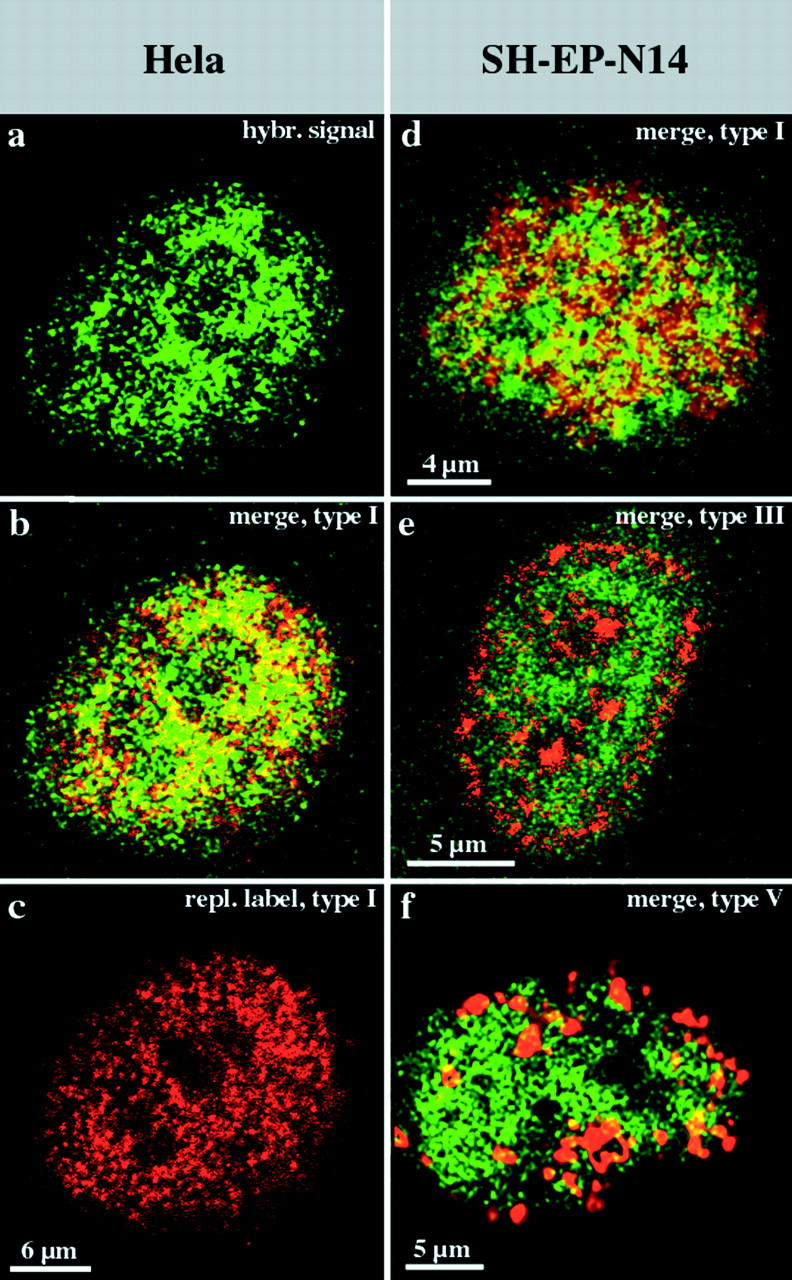
R-band DNA is confined to the interior compartment during interphase. Nuclei (a–c, HeLa S6 nucleus; and d–f, three different nuclei from SH-EP N14 cells) were replication-labeled with Cy3-dUTP (replication patterns depicted in red). DNA from the H3 isochore fraction (FITC-detected, depicted in green) was hybridized to Cy3-labeled nuclei. Nuclei were analyzed by confocal microscopy. For each nucleus, identical midnuclear planes are shown regarding FITC or Cy3 fluorescence detection (colocalizing FITC and Cy3 fluorescence appears yellow on merged [b and d–f] images). Regarding the HeLa nucleus, the merge (b) of the hybridization signal (a) and the type I replication pattern (c) reveals the localization of R-band sequences within the interior compartment. A similar localization of R-band sequences is obvious regarding SH-EP N14 cells (d, underlying type I pattern in red). R-band sequences are excluded from the peripheral (e, red, type III pattern) and late replicating (f, red, type V pattern) compartments. It should be noted that all confocal images presented were not further processed and reflect the resolution limits of light microscopy. Therefore, some colocalization of fluorescent signals at the boundaries of differently labeled distinct compartments is expected.
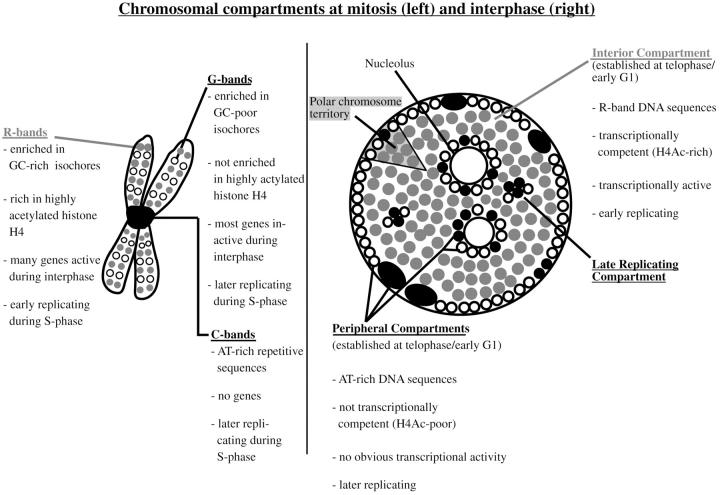
Figure 10.
The scheme summarizes previous results and the data of the present paper regarding compartmentalization of mammalian genomes during mitosis and interphase. The well characterized bands of mitotic chromosomes give rise to distinct higher order functional compartments within the cell nucleus. Distinct bands of mitotic chromosomes differ in a variety of features as isochore composition and corresponding DNA sequence composition (Bickmore and Sumner 1989; Craig and Bickmore 1993; Bernardi 1995), gene content (Bickmore and Sumner 1989; Craig and Bickmore 1993; Bernardi 1995; Cross et al. 1997), acetylation levels of histone H4 (Jeppesen and Turner 1993), transcriptional activity of genes (Craig and Bickmore 1993, Craig and Bickmore 1994), and replication timing during interphase (Dutrillaux et al. 1976; Camargo and Cervenka 1982). Differences in DNA sequence composition (Rae and Franke 1972; Manuelidis and Borden 1988; O'Keefe et al. 1992), acetylation levels of histone H4 (acetylated histone H4 designated as H4Ac), transcriptional activity, and replication timing of chromatin targeted to distinct nuclear compartments (Ferreira et al. 1997) demonstrate the functional features of these compartments and their relation to genome organization revealed by banding patterns of mitotic chromosomes. R-band sequences (symbolized by gray dots) localize to the interior compartment, whereas G- and C-band sequences localize to the peripheral and late replicating compartments (symbolized by black and open dots). In the present study, the peripheral and late replicating compartments revealed comparable properties. Higher order nuclear compartments are built up by chromosome territories displaying a distinct polarized distribution of R-band DNA and G/C-band DNA organized into corresponding subchromosomal foci (Zink et al. 1998, Zink et al. 1999).
Hyperacetylated Isoforms of Histone H4 Are Enriched within the Interior Compartment
As R-bands of mitotic chromosomes comprise most of the genes expressed during interphase, one would expect the transcriptionally competent chromatin within the interior compartment comprising the R-band sequences. Hyperacetylated isoforms of histone H4 are a characteristic feature of transcriptionally competent chromatin (Jeppesen 1996; Hassig and Schreiber 1997; Utley et al. 1998). To investigate whether transcriptionally competent chromatin is indeed confined to specific nuclear compartments, we stained replication-labeled nuclei with antiserum R 232/8 specifically recognizing hyperacetylated isoforms of histone H4 (see Materials and Methods).
Fig. 7 summarizes the results obtained for female HDFs (100 nuclei examined) and mouse myoblasts (70 nuclei examined). Unsynchronized, exponentially growing cultures were fixed 27 h after BrdU pulse labeling and immunostained. Comparison of BrdU labeling patterns and the nuclear distribution of hyperacetylated histone H4 isoforms revealed a clear correlation. Hyperacetylated histone H4 isoforms were detected throughout the whole interior compartment labeled by the type I pattern (Fig. 7, a–c and e), but were excluded from the peripheral and late replicating compartments (Fig. 7, f–h). Also, in female HDFs, a heterochromatic structure that presumably is the Barr body was not stained by antiserum R232/8 (Fig. 7b and Fig. d). This is in agreement with previous data demonstrating a lack of histone H4 acetylation for mammalian inactive X chromosomes during mitosis (Jeppesen and Turner 1993).
Figure 7.
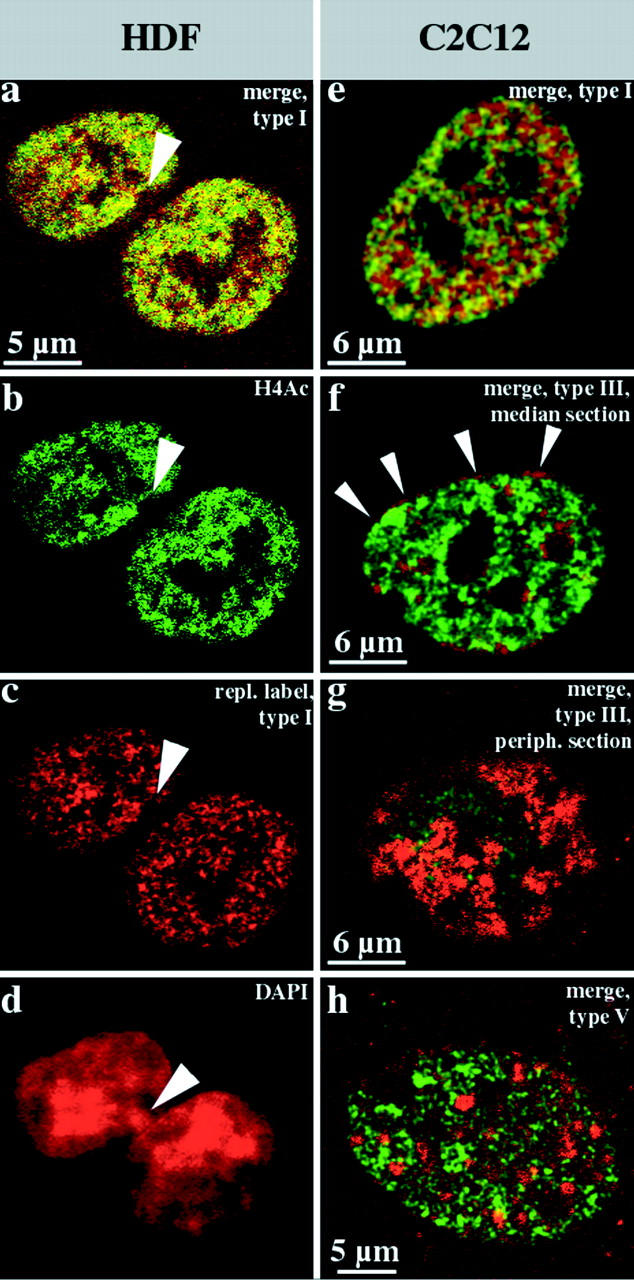
Hyperacetylated isoforms of histone H4 are confined to the interior compartment. Human diploid female fibroblasts (a–d, HDFs) and mouse C2C12 myoblasts (e–h) were replication-labeled with BrdU (TRITC-detected, depicted in red) for 30 min, fixed after 27 h, and immunostained with rabbit antiserum R232/8 specific for hyperacetylated histone H4 (H4Ac, FITC detected, depicted in green). a–c show light optical sections from identical midnuclear planes (b, FITC detection; c, TRITC detection; and a, merge, colocalizing FITC and TRITC signals appear yellow). Hyperacetylated histone H4 (b, FITC-detected, green) is confined to the interior compartment as the early replicating DNA (a and c, type I pattern, red). About 70% of these female HDFs display a strongly DAPI-stained domain at the nuclear periphery (arrowhead in d that is the corresponding epifluorescence DAPI image of the nucleus depicted in a–c). These domains, which likely represent the inactive X chromosome, are not immunostained by R232/8 antiserum and contain no early replicating DNA (a–c, arrowheads). The merges of single light optical sections detecting FITC (hyperacetylated histone H4, green) or TRITC (BrdU replication patterns, red) fluorescence (colocalizing FITC and TRITC signals appear yellow) of identical planes of corresponding C2C12 nuclei are shown in e–h. The nuclei display type I (e), type III (f and g), and type V (h) BrdU labeling patterns (red). Distinct focal planes of the same nucleus are depicted in f (midnuclear plane) and g (nuclear periphery). Note the concentration of R232/8 staining (green) within the interior compartment labeled by the type I pattern (e), whereas the peripheral (f and g) and late replicating (h) compartments are excluded. Note in particular, the absence of FITC fluorescence in the perinuclear region (the red rim around the nucleus in f shown by arrowheads and the red peripheral patches (segregated labeled and unlabeled territories) in g. The faint FITC signal in g is likely due to the low resolution of confocal microscopes along the optical axis.
As the transcriptional competence of genome compartments is an important feature with regard to their functional characteristics, we extended the analysis to CHO and HeLa S6 cells. Unsynchronized, exponentially growing cultures of HeLa or CHO cells were fixed 1 or 23 h after replication labeling with Cy3-dUTP to confirm similarity of results between S phase and non-S phase cells. Cy3-dUTP labeling excludes an influence of artificial changes of chromatin structure induced by DNA denaturation (Zink, D., unpublished results) that is necessary for BrdU detection on the results.
Cy3-dUTP–labeled CHO nuclei immunostained with R232/8 antiserum were examined and examples are shown in Fig. 8, a–i. For both groups (fixation after 1 h [n = 28] or 23 h [n = 30]) we obtained similar results. There was the same correlation between nuclear compartments revealed by replication labeling and the nuclear distribution of hyperacetylated histone H4 isoforms as observed for HDFs or C2C12 cells (compare Fig. 7 and Fig. 8). Hyperacetylated isoforms of histone H4 are confined to the interior compartment labeled by the type I pattern (Fig. 8, a–c). Type III–V patterns do not overlap with regions immunostained by antiserum R 232/8 (Fig. 8, d–i). Similar results were also obtained with HeLa S6 cells, fixed after 1 h (n = 20) or 23 h (n = 20) and double-labeled with Cy3-dUTP and antiserum R 232/8 (Fig. 8, j–l).
Figure 8.
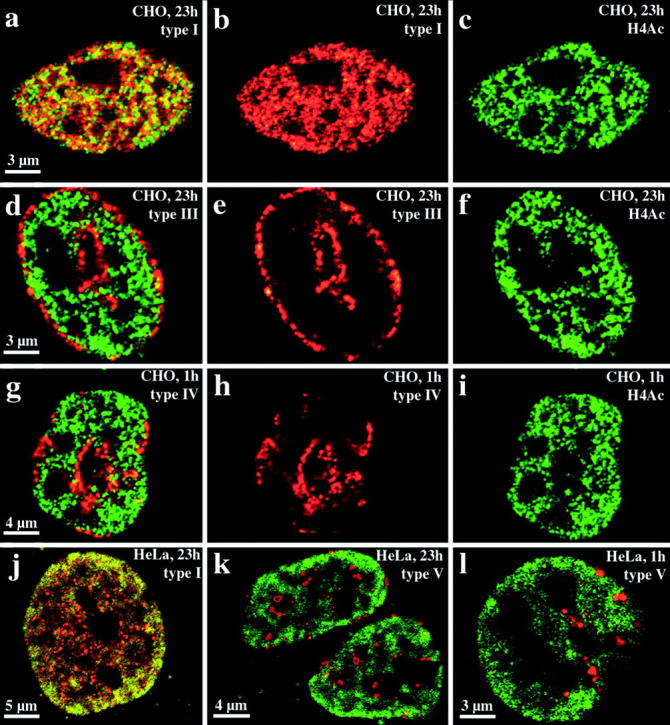
Compartmentaliza-tion of transcriptionally competent chromatin in nuclei of CHO and HeLa S6 cells. Nuclei were replication-labeled with Cy3-dUTP (depicted in red). Cells were fixed and immunostained with R232/8 antiserum (FITC-detected, depicted in green) 23 (a–f, j, and k) or 1 h (g–i, l) after replication labeling. a–i display images of CHO nuclei, whereas j–l show HeLa S6 nuclei. For each nucleus, single light optical sections of identical midnuclear planes regarding Cy3 and FITC fluorescence detection are shown. For all distinct nuclei (a–c, d–f, g–i, and j and k or l correspond to one nucleus) the FITC (c, f, and i), Cy3 (b, e, and h) and merged signals (a, d, and g) are shown except for j–l where only the merged signals of different nuclei are depicted. Colocalizing FITC and Cy3 signals appear yellow on merged images. Chromatin enriched in highly acetylated isoforms of histone H4 (H4Ac), indicated by R232/8 staining (green), is concentrated in the interior compartment (a, b, and j, colocalizing with the type I pattern, red). R232/8 staining is excluded from the peripheral and late replicating compartments (d, e, g, h, k, and l, replication-labeled, red).
Transcriptional Activity Is Compartmentalized within Mammalian Interphase Nuclei
The results suggested that not only transcriptional competence, but also that the process of transcription itself also might be subject of higher order nuclear compartmentalization. Nuclear higher order compartments were visualized within HeLa S6 and CHO nuclei by replication labeling with FITC-dUTP (synchronized and unsynchronized cultures used). On the next day, FITC-labeled cells were microinjected with BrUTP (incorporated into nascent RNA; Wansink et al. 1993) and fixed after 10 min of microinjection. Immunostaining of incorporated BrUTP revealed only the interior compartment as the transcriptionally active compartment. (Fig. 9, a–c). No considerable BrUTP labeling was visible within the peripheral compartments (Fig. 9, d–f). There was also no overlap between type IV and V FITC patterns and BrUTP incorporation. Similar results were obtained for HeLa and CHO cells. Therefore, the interior compartment comprising the early replicating R-band sequences harbors the transcriptionally competent as well as the actively transcribed chromatin. The data are summarized in Fig. 10.
Figure 9.
Transcriptional activity is confined to the interior compartment. Transcriptional activity within distinct genome compartments was investigated by BrUTP pulse labeling (10 min) of cells replication-labeled on the previous day. BrUTP was detected with TRITC fluorescence, whereas replication labeling was performed with FITC-dUTP. HeLa cells (a–c) and CHO cells (d–f) were investigated. Identical nuclear planes were imaged with regard to FITC detection (b and e, depicted in green) and TRITC detection (c and f, depicted in red). Merges of corresponding FITC and TRITC signals are depicted in a and d (colocalizing signals appear yellow). Transcriptional activity, indicated by BrUTP incorporation into nascent RNA (c and f, and, a and d, red) is confined to the interior compartment (a and b, green, colocalizing with the type I pattern). The TRITC signal in the upper left regions of a and c is due to an adjacent BrUTP-labeled nucleus that is not replication-labeled. d–f depict the exclusion of RNA synthesis from the peripheral compartments (d and e, green). Arrows in f show examples of perinuclear and perinucleolar regions strongly labeled by the type III replication labeling pattern (d and e, green) that display no TRITC signal (no RNA synthesis).
Discussion
In this study, we characterized the functional compartmentalization of mammalian genomes during interphase. The data demonstrated that a specific pattern of spatial genome compartmentalization was present during all interphase stages and was clonally inherited. Distinct compartments comprised DNA sequences belonging to distinct chromosomal bands during mitosis. R-band sequences built up a coherent compartment within the nuclear interior, whereas G/C-band sequences localize to compartments at the nuclear and nucleolar peripheries as well as to minor internal compartments. The early replicating compartment comprising the R-band sequences harbored the transcriptionally competent chromatin. Detectable nascent RNA synthesis was confined to the interior compartment. Fig. 10 summarizes the correlations between functional higher order nuclear genome architecture and chromosome organization during mitosis and interphase.
The functionally different higher order compartments were built up through the alignment of polar chromosome territories with clusters of early replicating DNA directed towards the nuclear interior and clusters of later replicating DNA located at the nuclear or nucleolar peripheries. We demonstrated recently that early replicating R- or later replicating G/C-bands of mitotic chromosomes are retained as distinct chromosomal domains termed subchromosomal foci (SF) within interphase chromosome territories (Zink et al. 1998, Zink et al. 1999). Within chromosome territories of cycling (G1) cells, we observed a clustering of R-SF or G/C-SF at distinct sites of the territories (Bornfleth et al. 1999; Zink et al. 1999). Therefore, the emerging picture is as follows: distinct bands (in the megabase pair size range) alternating on mitotic chromosomes are retained as distinct domains during interphase (SF), but are reorganized within the territories. The distinct SF cluster within a territory and, therefore, give rise to a polar organization of this structure in the size range of several tens to hundreds megabase pairs (Morton 1991). Alignment of these polar territories creates higher order functional genome compartments (size range: gigabase pairs) within mammalian cell nuclei (see also Ferreira et al. 1997; Lamond and Earnshaw 1998).
The results and conclusions of the present study are in agreement with the present understanding of chromosome organization on the one hand, and a growing body of evidence indicating functional higher order nuclear compartmentalization on the other. The fact that R-band sequences localize within the compartment harboring the transcriptionally competent and active parts of the genome is in agreement with the fact that R-bands contain the majority of genes and in particular those genes that are actively transcribed during interphase (Bickmore and Sumner 1989; Craig and Bickmore 1993, Craig and Bickmore 1994; Cross et al. 1997). Sequence identity was confirmed by in situ hybridization with DNA from the H3 isochore fraction (Saccone et al. 1996). Under the conditions we used, this probe hybridized specifically to sequences present within most of the R-bands of the human karyotype. Although previous studies described hybridization only to sequences present within a subset of R-bands (Saccone et al. 1996), the result may strongly depend on the actual hybridization conditions. Hybridization to metaphase spreads of human lymphocytes and neuroblastoma cells and the reproducible R-banding observed on chromosomes, demonstrated that the H3 isochore DNA fraction is a reliable probe for R-band DNA under the hybridization conditions we used. Thus, the data clearly demonstrated that R-band sequences localize to the interior compartment.
However, the in situ hybridization data do not rule out that these sequences also contribute to other compartments. We presently cannot definitely exclude that a lack of signal in unlabeled compartments is due to accessibility problems. However, we think this is unlikely as we characterized R-band chromatin not only by sequence identity, but also by its replication timing and its content of hyperacetylated histone H4. Early replicating sequences do not obviously contribute to peripheral and late replicating compartments that are also not enriched in hyperacetylated histone H4. As both features are characteristic for R-band chromatin (Camargo and Cervenka 1982; Jeppesen and Turner 1993), together the data indicate that R-band chromatin does not make a major contribution to peripheral and late replicating compartments.
Although we did not use specific DNA probes to localize G- and C-band DNA within the nucleus, a variety of data indicate that the corresponding sequences localize in the peripheral and late replicating compartments. First, many studies describe the detection of C-band sequences at the corresponding nuclear positions (Rae and Franke 1972; Manuelidis and Borden 1988; O'Keefe et al. 1992), whereas the localization of G-band sequences at these nuclear positions was suggested by replication labeling studies (Ferreira et al. 1997). G/C-band sequences are known to replicate during the second half of S phase (Kim et al. 1975; Dutrillaux et al. 1976; Camargo and Cervenka 1982; O'Keefe et al. 1992; Ferreira et al. 1997). As our results show that DNA within the interior compartment replicates early in S phase, G- or C-band sequences cannot make a major contribution to this compartment and, therefore, have to be confined to the nuclear compartments comprising the transcriptionally inactive parts of the genome. This is in agreement with the fact that G-band DNA displays a low density of genes that are mostly not expressed and the fact that C-bands comprise highly repetitive sequences (Goldman et al. 1984; Bickmore and Sumner 1989; Craig and Bickmore 1993).
The data are in agreement with many studies indicating that expressed sequences are generally located within the nuclear interior, whereas repressed sequences locate towards the nuclear periphery often in close association with constitutive heterochromatin (C-bands of mammalian chromosomes comprise the constitutive heterochromatin) (Hoehn and Martin 1973; Mathog et al. 1984; Belmont et al. 1986; Manuelidis and Borden 1988; Csink and Henikoff 1996; Maillet et al. 1996; Brown et al. 1997; Imai et al. 1997; Andrulis et al. 1998). However, these studies were performed with a variety of eukaryotic taxa as different as yeast, flies, and mammals. Although the principle of compartmentalization with repressed sequences at perinuclear positions might be general, experimental data indicate that the type of DNA sequences and chromosomal structures located within the different nuclear compartments, as well as the mechanisms mediating genome compartmentalization, might be different in distinct taxa (Rabl 1885; Hochstrasser et al. 1986; Haaf and Schmid 1991; Manuelidis and Borden 1988; Palladino et al. 1993).
With regard to the specific localization of chromatin observed in mammals, previous analyses indicated that the process of transcription might not be responsible (Ferreira et al. 1997) for nuclear genome compartmentalization in mammals. The fact that the nuclear compartmentalization of mammalian genomes is strongly related to the banding patterns of mitotic chromosomes, and that the nuclear compartmentalization is established immediately after mitosis imply that chromatin belonging to the different bands is targeted to distinct nuclear positions when the nucleus reconstitutes. This process might take place independently for each chromosome (Ferreira et al. 1997). In this case, DNA or chromatin belonging to distinct bands of a chromosome has to be specifically recognized. Recognition could take place at the level of DNA sequence as R- and G/C-bands display a distinct isochore composition (Bernardi 1995). However, we think that unlikely as the active (Xa) and inactive (Xi) X chromosomes of female mammals display a similar DNA sequence, but are affected by the nuclear genome compartmentalization regarding transcriptional competence and activity (Xi is usually located at the nuclear periphery; Hoehn and Martin 1973; Belmont et al. 1986). As there are specific modifications of R- or G/C-band chromatin (Jeppesen and Turner 1993; Haaf 1995) that affect also Xa and Xi (Xi is exceptional in the sense that chromatin modifications affect almost the whole chromosome rather than its distinct bands), specific recognition at this level seems possible.
Although some chromatin domains might display dynamic relocalizations according to changes in their functional states, the stable maintenance of replication labeling patterns indicates that positional changes of chromosomes or chromosomal regions might be exceptional events within the nucleus. This is in full agreement with recent studies of DNA dynamics within nuclei of living mammalian cells (Robinett et al. 1996; Shelby et al. 1996; Abney et al. 1997; Marshall et al. 1997b; Zink and Cremer 1998; Zink et al. 1998). Nevertheless, rare large-scale movements of chromosomes or parts of them were observed in living cell studies (Shelby et al. 1996; Zink and Cremer 1998; Zink et al. 1998). Such movements are compatible with the observed, stable compartmentalization of functionally defined chromatin, as long as they occur within or between corresponding compartments. Whereas the intranuclear location of the compartments is fixed, at least within similar cell types, there may be a degree of flexibility in the positioning of individual DNA sequences within the compartments. For example, if a centromeric C-band domain moves during interphase from the nucleolar to the nuclear periphery, the overall compartmentalization is not disturbed. In this regard, the observed stable compartmentalization is also compatible with the dynamic repositioning of nuclear domains observed in fixed cell studies (for review see De Boni 1994).
Acknowledgments
We thank Irina Solovei for advice regarding H3 isochore probe hybridization, Roel van Driel and Pernette Verschure for advice in BrUTP labeling, and Robert Martin for help in arrangement of images.
This work was supported by grants from the Wellcome Trust (045030/Z/95 to B.M. Turner) and the Deutsche Forschungsgemeinschaft (Zi 560/2-1).
Footnotes
1.used in this paper: BrdU, bromodeoxyuridine; CldU, chlorodeoxyuridine; DAPI, 4′,6-diamidino-2-phenylindole; HDF, human diploid fibroblast; IdU, iododeoxyuridine; SF, subchromosomal foci
References
- Abney J.R., Cutler B., Fillbach M.L., Axelrod D., Scalettar B.A. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J. Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Neimann A.M., Zappulla D.C., Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Belmont A.S., Bignone F., Ts'o P.O.P. The relative intranuclear positions of Barr bodies in XXX non-transformed human fibroblasts. Exp. Cell Res. 1986;165:165–179. doi: 10.1016/0014-4827(86)90541-0. [DOI] [PubMed] [Google Scholar]
- Belmont A.S., Bruce K. Visualization of G1 chromosomesa folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R., Mortillaro M.J., Ma H., Wei X., Samarabandu J. The nuclear matrixa structural milieu for genomic function. Int. Rev. Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome. Annu. Rev. Genet. 1989;23:637–661. doi: 10.1146/annurev.ge.23.120189.003225. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The human genomeorganization and evolutionary history. Annu. Rev. Genet. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985;228:953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bickmore W.A., Sumner A.T. Mammalian chromosome banding—an expression of genome organization. Trends Genet. 1989;5:144–148. doi: 10.1016/0168-9525(89)90055-3. [DOI] [PubMed] [Google Scholar]
- Blobel G. Gene gatinga hypothesis. Proc. Natl. Acad. Sci. USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden J., Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988;242:1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- Bornfleth H., Edelmann P., Zink D., Cremer C. Three-dimensional analysis of genome topology. In: Jähne B., Haubecker H., Geibler P., editors. Handbook of Computer Visions and Applications. Vol 3. Academic Press Inc; San Diego/New York: 1999. pp. 859–878. [Google Scholar]
- Bridger J.M., Bickmore W.A. Putting the genome on the map. Trends Genet. 1998;14:403–409. doi: 10.1016/s0168-9525(98)01572-8. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M., Fisher A.G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Camargo M., Cervenka J. Patterns of DNA replication of human chromosomes. II. Replication map and replication model. Am. J. Hum. Genet. 1982;34:757–780. [PMC free article] [PubMed] [Google Scholar]
- Craig J.M., Bickmore W.A. Chromosome bands—flavours to savour. Bioessays. 1993;15:349–354. doi: 10.1002/bies.950150510. [DOI] [PubMed] [Google Scholar]
- Craig J.M., Bickmore W.A. The distribution of CpG islands in mammalian chromosomes. Nat. Genet. 1994;7:376–382. doi: 10.1038/ng0794-376. [DOI] [PubMed] [Google Scholar]
- Cremer T., Lichter P., Borden J., Ward D.C., Manuelidis L. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome-specific library probes. Hum. Genet. 1988;80:235–246. doi: 10.1007/BF01790091. [DOI] [PubMed] [Google Scholar]
- Cremer T., Dietzel S., Eils R., Lichter P., Cremer C. Chromosome territories, nuclear matrix filaments and inter-chromatin channelsa topological view on nuclear architecture and function. In: Brandham P.E., Bennett M.D., editors. Kew Chromosome Conference IV. Royal Botanic Gardens; Kew, United Kingdom: 1995. pp. 63–81. [Google Scholar]
- Cross S.H., Lee M., Clark V.H., Craig J.M., Bird A.P., Bickmore W.A. The chromosomal distribution of CpG islands in the mouseevidence for genome scrambling in the rodent lineage. Genomics. 1997;40:454–461. doi: 10.1006/geno.1996.4598. [DOI] [PubMed] [Google Scholar]
- Csink A., Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila . Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- Cuny G., Soriano P., Macaya G., Bernardi G. The major components of the mouse and human genomespreparation, basic properties and compositional heterogeneity. Eur. J. Biochem. 1981;111:227–233. doi: 10.1111/j.1432-1033.1981.tb05227.x. [DOI] [PubMed] [Google Scholar]
- De Boni U. The interphase nucleus as a dynamic structure. Int. Rev. Cytol. 1994;150:149–171. doi: 10.1016/s0074-7696(08)61541-7. [DOI] [PubMed] [Google Scholar]
- Dutrillaux B., Couturier J., Richer C.-L., Viegas-Peguinot E. Sequence of DNA replication in 277 R- and Q-bands of human chromosomes using a BrdU treatment. Chromosoma. 1976;58:51–61. doi: 10.1007/BF00293440. [DOI] [PubMed] [Google Scholar]
- Eils R., Dietzel S., Bertin E., Schroeck E., Speicher M.R., Ried T., Robert-Nicoud M., Cremer C., Cremer T. Three-dimensional reconstruction of painted human interphase chromosomesactive and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J. Cell Biol. 1996;135:1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauth C. Evaluierung eines verfahrens zur darstellung von chromosomen in lebenden säugetierzellen. Diploma thesis 1998. Institute of Anthropology and Human Genetics; LMU München, Germany: pp. 1–102 [Google Scholar]
- Federico C., Saccone S., Bernardi G. The gene-richest bands of human chromosomes replicate at the onset of the S phase. Cytogenet. Cell Genet. 1998;80:83–88. doi: 10.1159/000014961. [DOI] [PubMed] [Google Scholar]
- Ferreira J., Paolella G., Ramos C., Lamond A.I. Spatial organization of large-scale chromatin domains in the nucleusa magnified view of single chromosome territories. J. Cell Biol. 1997;139:1597–1610. doi: 10.1083/jcb.139.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M.A., Holmquist G.P., Gray M.C., Caston L.A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Haaf T. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and functionimplications for methylation-associated cellular processes. Pharmacol. Ther. 1995;65:19–46. doi: 10.1016/0163-7258(94)00053-6. [DOI] [PubMed] [Google Scholar]
- Haaf T., Schmid M. Chromosome topology in mammalian interphase nuclei. Exp. Cell Res. 1991;192:325–332. doi: 10.1016/0014-4827(91)90048-y. [DOI] [PubMed] [Google Scholar]
- Hassig C.A., Schreiber S.L. Nuclear histone acetylases and deacetylases and transcriptional regulationHATs off to HDACs. Curr. Opin. Chem. Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M., Sedat J. Three-dimensional organization of Drosophila melanogaster interphase nuclei. II. Chromosome spatial organization and gene regulation. J. Cell Biol. 1987;104:1471–1483. doi: 10.1083/jcb.104.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M., Mathog D., Gruenbaum Y., Saumweber H., Sedat J.W. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster . J. Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn H., Martin G.M. Nonrandom arrangement of human chromatintopography of disomic markers X, Y, and 1h plus 1. Cytogenet. Cell Genet. 1973;12:443–452. doi: 10.1159/000130487. [DOI] [PubMed] [Google Scholar]
- Hutchinson N., Weintraub H. Localization of DNAse I-sensitive sequences to specific regions of interphase nuclei. Cell. 1985;43:471–482. doi: 10.1016/0092-8674(85)90177-1. [DOI] [PubMed] [Google Scholar]
- Imai S., Nishibayashi S., Takao K., Tomifuji M., Fujino T., Hasegawa M., Takano T. Dissociation of Oct-1 from the nuclear peripheral structure induces the cellular aging-associated collagenase gene expression. Mol. Biol. Cell. 1997;8:2407–2419. doi: 10.1091/mbc.8.12.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Pombo A. Replicon clusters are stable units of chromosome structureevidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P. Histone acetylationa possible mechanism for the inheritance of cell memory at mitosis. Bioessays. 1996;19:67–74. doi: 10.1002/bies.950190111. [DOI] [PubMed] [Google Scholar]
- Jeppesen P., Turner B.M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Kim M.A., Johannsmann R., Grzeschik K.-H. Giemsa staining of the sites replicating DNA early in human lymphocyte chromosomes. Cytogenet. Cell Genet. 1975;15:363–371. doi: 10.1159/000130535. [DOI] [PubMed] [Google Scholar]
- Kurz A., Lampel S., Nickolenko J.E., Bradl J., Benner A., Zirbel R.M., Cremer T., Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 1996;135:1195–1202. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I., Earnshaw W.C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Leitch A.R., Mosgoller W., Schwarzacher T., Bennett M.D., Heslop-Harrison J.S. Genomic in situ hybridization to sectioned nuclei shows chromosome domains in grass hybrids. J. Cell Sci. 1990;95:335–341. doi: 10.1242/jcs.95.3.335. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Cardoso C. Targeting and association of proteins with functional domains in the nucleusthe insoluble solution. Int. Rev. Cytol. 1995;162B:303–335. doi: 10.1016/s0074-7696(08)62620-0. [DOI] [PubMed] [Google Scholar]
- Lichter P., Cremer T., Borden J., Manuelidis L., Ward D.C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Ma H., Samarabandu J., Devdhar R.S., Acharya R., Cheng P.-C., Meng C., Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor H., Varley J. Working with Animal Chromosomes 1988. John Wiley & Sons, Inc; Chichester, New York/Brisbane, Toronto/Singapore: pp. 290 [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., Gasser S.M. Evidence for silencing compartments within the yeast nucleusa role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Individual interphase chromosome domains revealed by in situ hybridization. Hum. Genet. 1985;71:288–293. doi: 10.1007/BF00388453. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A view of interphase chromosomes. Science. 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Borden J. Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situ hybridization and three-dimensional reconstruction. Chromosoma. 1988;96:397–410. doi: 10.1007/BF00303033. [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Fung J.C., Sedat J.W. Deconstructing the nucleusglobal architecture from local interactions Curr. Opin. Genet. Dev 7 1997. 259 263a [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Straight A., Marko J.F., Swedlow J., Dernburg A., Belmont A., Murray A.W., Agard D.A., Sedat J.W. Interphase chromosomes undergo constrained diffusional motion in living cells Curr. Biol 7 1997. 930 939b [DOI] [PubMed] [Google Scholar]
- Mathog D., Hochstrasser M., Gruenbaum Y., Saumweber H., Sedat J. Characteristic folding pattern of polytene chromosomes in Drosophila salivary gland nuclei. Nature. 1984;308:414–421. doi: 10.1038/308414a0. [DOI] [PubMed] [Google Scholar]
- Morton N.E. Parameters of the human genome. Proc. Natl. Acad. Sci. USA. 1991;88:7474–7476. doi: 10.1073/pnas.88.17.7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu H., Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe R.T., Henderson S.C., Spector D.L. Dynamic organization of DNA replication in mammalian cell nucleispatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., Gasser S.M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- Pyrpasopoulou A., Meier J., Maison C., Simos G., Georgatos S.D. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:7108–7119. [PMC free article] [PubMed] [Google Scholar]
- Rabl, C. 1885. Ueber Zelltheilung. Morphol. Jahrbuch. 10.
- Rae P.M.M., Franke W.W. The interphase distribution of satellite DNA-containing heterochromatin in mouse nuclei. Chromosoma. 1972;39:443–456. doi: 10.1007/BF00326177. [DOI] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Wilhelm C., Sudlow G., Murray A., Belmont A.S. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross R.A., Spengler B.A., Biedler J.L. Coordinate morphological and biochemical interconversion of human neuroblastoma cells. J. Natl. Cancer Inst. 1983;71:741–747. [PubMed] [Google Scholar]
- Saccone S., De Sario A., Wiegant J., Raap A.K., Della Valle G., Bernardi G. Correlations between isochores and chromosomal bands in the human genome. Proc. Natl. Acad. Sci. USA. 1993;90:11929–11933. doi: 10.1073/pnas.90.24.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S., Caccio S., Kusuda J., Andreozzi L., Bernardi G. Identification of the gene-richest bands in human chromosomes. Gene. 1996;174:85–94. doi: 10.1016/0378-1119(96)00392-7. [DOI] [PubMed] [Google Scholar]
- Schardin M., Cremer T., Hager H.D., Lang M. Specific staining of human chromosome position in chinese hamster × man hybrid cell lines demonstrates interphase chromosome territories. Hum. Genet. 1985;71:281–287. doi: 10.1007/BF00388452. [DOI] [PubMed] [Google Scholar]
- Shelby R.D., Hahn K.M., Sullivan K.F. Dynamic elastic behavior of α-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer H., Green M.R. Compartmentalization of eukaryotic gene expressioncauses and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- Sparvoli E., Levi M., Rossi E. Replication clusters may form structurally stable complexes of chromatin and chromosomes. J. Cell Sci. 1994;107:3097–3103. doi: 10.1242/jcs.107.11.3097. [DOI] [PubMed] [Google Scholar]
- Spector D.L. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Strouboulis J., Wolffe A.P. Functional compartmentalization of the nucleus. J. Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- Taylor J.A. A Brief History of the Discovery of Sister Chromatid Exchanges 1984. Plenum Press; New York/London: pp. 1–10 [DOI] [PubMed] [Google Scholar]
- Telenius H., Carter N.P., Bepp C.E., Nordenskjöld M., Ponder B.A.J., Tunnacliffe A. Degenerate oligonucleotide-primed PCRgeneral amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Utley R.T., Ikeda K., Grant P.A., Cote J., Steger D.J., Eberharter A., John S., Workman J.L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- van Driel R., Wansink D.G., van Steensel B., Grande M.A., Schul W., de Jong L. Nuclear domains and the nuclear matrix. Int. Rev. Cytol. 1995;162A:151–189. doi: 10.1016/s0074-7696(08)61231-0. [DOI] [PubMed] [Google Scholar]
- Wansink D.G., Schul W., van der Kraan I., van Steensel B., van Driel R., de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II domains scattetred throughout the nucleus. J. Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A., Cziepluch C., Hamann U., Schuermann J., Schwab M. The N-Myc oncoprotein is associated in vivo with the phosphoprotein Max(p20/22) in human neuroblastoma cells. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:3703–3712. doi: 10.1002/j.1460-2075.1991.tb04938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D., Cremer T. Chromosome dynamics in nuclei of living cells. Curr. Biol. 1998;8:R321–R324. doi: 10.1016/s0960-9822(98)00198-5. [DOI] [PubMed] [Google Scholar]
- Zink D., Cremer T., Saffrich R., Fischer R., Trendelenburg M.F., Ansorge W., Stelzer E.H.K. Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]
- Zink D., Bornfleth H., Visser A., Cremer C., Cremer T. Organization of early and late replicating DNA in human chromosome territories. Exp. Cell Res. 1999;247:176–188. doi: 10.1006/excr.1998.4311. [DOI] [PubMed] [Google Scholar]