Abstract
Cells adhere to the substratum through specialized structures that are linked to the actin cytoskeleton. Recent studies report that adhesion also involves the intermediate filament (IF) and microtubule cytoskeletons, although their mechanisms of interaction are unknown. Here we report evidence for a novel adhesion-dependent interaction between components of the actin and IF cytoskeletons. In biochemical fractionation experiments, fimbrin and vimentin coprecipitate from detergent extracts of macrophages using vimentin- or fimbrin-specific antisera. Fluorescence microscopy confirms the biochemical association. Both proteins colocalized to podosomes in the earliest stages of cell adhesion and spreading. The complex is also found in filopodia and retraction fibers. After detergent extraction, fimbrin and vimentin staining of podosomes, filopodia, and retraction fibers are lost, confirming that the complex is localized to these structures. A 1:4 stoichiometry of fimbrin binding to vimentin and a low percentage (1%) of the extracted vimentin suggest that fimbrin interacts with a vimentin subunit. A fimbrin-binding site was identified in the NH2-terminal domain of vimentin and the vimentin binding site at residues 143–188 in the CH1 domain of fimbrin. Based on these observations, we propose that a fimbrin–vimentin complex may be involved in directing the assembly of the vimentin cytoskeleton at cell adhesion sites.
Keywords: fimbrin, vimentin, cytoskeleton, adhesion, microfilaments
The intricate structure of the microfilament (MF),1 microtubule (MT), and intermediate filament (IF) cytoskeletons is organized by an assortment of accessory proteins that stabilize or cross-link filaments or attach filaments to the membranes of organelles and the cell surface (Schliwa 1986; Bershadsky and Vasiliev 1988). In most cases, we know much about the function of the binding proteins within one cytoskeletal system; for example, the role of actin cross-linking proteins in bundles and networks of actin filaments (Matsudaira 1991, Matsudaira 1994). In contrast, relatively little is known whether or how the coassembly and coorganization of cytoskeletal systems are integrated and coordinated. The very earliest work used MT and IF depolymerizing drugs to demonstrate that the organization of the two cytoskeletal systems are interdependent; the disassembly of one disrupts the pattern of the other (Goldman and Knipe 1972; Croop and Holtzer 1975; Blose and Chacko 1976; Wang and Choppin 1981). More recent studies have identified several proteins including the IF-associated protein, plectin, that could serve as physical cross-links between the different cytoskeletal systems (Svitkina et al. 1996). In addition to biochemical demonstration of binding, electron micrographs of cultured fibroblast cells show plectin molecules bridging MTs and vimentin filaments. However, the story is likely to be more complicated because plectin is also implicated in interactions with the actin cytoskeleton based on the presence of an actin-binding domain (ABD) in the protein sequence (Wiche et al. 1991, Wiche et al. 1993; Liu et al. 1996). Thus, it is possible that cytoskeletal-binding proteins may interact with several cytoskeletal systems.
An emerging model is that a superfamily of actin-binding proteins containing a calponin homology (CH) domain may play an important integrative function in controlling the organization of the major cytoskeletal systems in the cell. In most cases, this CH domain is duplicated to form the classic ABD of fimbrin, α-actinin, spectrin, filamin, dystrophin, other actin cross-linking proteins, and the IF-binding protein, plectin (Dubreuil 1991; Hartwig and Kwiatkowski 1991; Otto 1994). In contrast, a single CH domain is found at the NH2 terminus of small G-protein regulatory molecules, vav and IQ GAP (Castresana and Saraste 1995; Brill et al. 1996). The list of functions attributed to these proteins implicate the CH domain in cross-linking MFs into bundles and networks and tethering IF and signaling proteins to the actin cytoskeleton (Castresana and Saraste 1995). Thus, CH domain proteins may directly control the structural organization of the cytoskeleton through one or more signaling pathways.
The experiments reported in this paper describe a new type of interaction between a subunit of the vimentin IF and an actin cross-linking protein, fimbrin. Fimbrin is a 68-kD actin cross-linking protein associated with actin bundles (Matsudaira and Burgess 1979; Bretscher and Weber 1980) in microvilli on the cell surface and in membrane ruffles, microspikes, and cell adhesion sites at the cell–matrix interface (Bretscher and Weber 1980; Marchisio et al. 1987; Matsudaira 1994). Conserved in sequence from yeast to humans, fimbrin consists of an NH2-terminal 12-kD calcium-binding domain followed by a pair of 27-kD actin-binding domains (ABD1 and ABD2) (Matsudaira 1991). Based on sequence and structure from x-ray crystallography, the ABDs consist of a pair of CH domains (de Arruda et al. 1990; Goldsmith et al. 1997). In mammals, three fimbrin isoforms are expressed in a tissue-specific pattern. The intestine-specific isoform, I-fimbrin, is localized to the brush border of intestinal and kidney epithelia (Lin et al. 1994). In contrast, the leukocyte-specific isoform, L-fimbrin, and the general isoform, T-fimbrin, are localized to cell–substratum adhesion sites in macrophages and fibroblasts and to the basal surface of differentiating intestinal enterocytes (Lin et al. 1993; Chafel et al. 1995). The adhesion site localization of fimbrin is curious because it does not label the actin bundle of the stress fiber but instead the site where the bundle is associated with cell–matrix adhesion proteins. This location at the base of the cell suggests a different function for the protein, possibly as a component of a cell adhesion complex involved in establishing cell polarity during embryogenesis. An important clue to the mechanism on how fimbrin is targeted to the base of the cell may be revealed by the identity of proteins that coprecipitated with fimbrin in adherent macrophages (Messier et al. 1993). The results in this paper show that fimbrin is complexed with vimentin subunits rather than vimentin filaments and both proteins colocalize at filopodia, retraction fibers, and podosomes on the ventral surface of cultured macrophages. The association between the two proteins is adhesion-dependent, implicating a mechanism that coregulates assembly of the two cytoskeletal systems at sites of cell adhesion.
Materials and Methods
Reagents
Unless stated otherwise, cell culture media and supplements were obtained from GIBCO BRL and reagents were obtained from Sigma Chemical Co.
Recombinant Proteins
Recombinant vimentin was expressed and purified by published protocols (Chou et al. 1997). Mutagenesis was carried out on cloned human vimentin in the pTZ18U vector (Muta-Gene Phagemid In Vitro Mutagenesis kit; Bio-Rad Laboratories). The 102C vimentin clone was generated by the deletion of the sequence encoding the NH2 terminus of vimentin and by the introduction of a new initiation ATG codon immediately preceding the codon for asparagine-102. For the N410 vimentin clone, a stop codon was introduced after the codon for isoleucine 410. Both constructs were subcloned into the PET-7 vector for expression in Escherichia coli.
For fimbrin expression and purification, BL21(DE3) cells were transformed and the cells were plated onto ampicillin plates. Colonies were inoculated into 2 liters of 2× YT media and shaken for 24 h at 37°C. The cells were harvested by centrifugation (4,000 rpm for 30 min) and suspended in 50 mM Tris, pH 8.0, 1 mM EDTA, 2 mM DTT, 1 mM PMSF, and 25% sucrose, and frozen. The cell suspension was thawed, lysed by sonication, and centrifuged at 18,000 rpm for 30 min. The supernatant was dialyzed into 10 mM Tris, pH 8.0, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 1 mM sodium azide and loaded onto a DEAE-Sepharose fast flow column (Pharmacia Biotech) preequilibrated with the same buffer. After washing to remove unbound material, the column was eluted with a linear gradient of NaCl (0–300 mM). Peak fractions were pooled, concentrated, and loaded onto a Q-Sepharose column (Pharmacia Biotech) that was equilibrated in 10 mM Pipes, pH 7.0, 1 mM EDTA, 50 mM NaCl, 1 mM DTT, 0.1 mM PMSF, and 0.1 mM sodium azide. The protein was again eluted with a linear salt gradient (0–300 mM NaCl), pooled, and loaded onto a Sephacryl S200 HR gel filtration column (Pharmacia Biotech). The purified protein was concentrated on a microconcentrator (30-kD cut-off).
Glutathione-S-transferase (GST) fusion constructs were generated by PCR cloning of human L-fimbrin. cDNA regions corresponding to L-fimbrin protein sequences 97-248, 243-385, 97-188, 97-142, 143-188, and 119-160 were amplified using synthetic oligonucleotide primers (Life Technologies, Inc.). A nucleotide tail coding for an in-frame BamHI restriction site was engineered into each 5′ PCR oligo; a nucleotide tail coding for an EcoRI restriction site and a downstream amber stop codon (TAG) was engineered into each 3′ PCR oligo. PCR-amplified DNAs were subcloned, via the engineered BamHI and EcoRI restriction sites, into a GST-vector, pGEX-4T2 (Pharmacia Biotech), directly downstream of the GST transcription region. GST-fimbrin fusion proteins were produced by inducing log phase DH5-α bacteria containing the GST construct with isopropyl-β-d-thiogalactopyranoside for 1.5 h, purifying the recombinant protein with glutathione agarose beads, and concentrating the eluate with a centriprep/centricon concentrator (Amicon).
Cell Culture
The cells used in these experiments were mouse macrophage cell lines P388D1 (ATCC TIB-63) and IC-21 (ATCC TIB-186). The cell lines were cultured in 85% RPMI-1640 medium, 15% FBS, and antibiotics (50 IU penicillin and 50 μg/ml streptomycin). Cell culture and maintenance techniques were performed as described by American Type Culture Collection.
Cell Extraction and Immunoprecipitation
P388D1 cells were grown to confluence (adherent cells), washed once with PBS, scraped off the plate in PBS, and pelleted by centrifugation. P388D1 cells were also grown in suspension (nonadherent cells), washed twice in PBS, and pelleted by centrifugation. The cells were lysed on ice in 10 mM Pipes buffer, pH 6.8, containing 0.5% Triton X-100, 300 mM sucrose, 100 mM KCl, 3 mM MgCl2, 10 mM EGTA, 2 mM PMSF, and 50 μM sodium vanadate (Messier et al. 1993). After 3 min, the cells were centrifuged at 10,000 rpm for 10 min. Aliquots of the supernatant representing the Triton-extractable fraction were precleared with protein A conjugated to agarose beads. For immunoprecipitation of fimbrin, the precleared supernatants were incubated with different antifimbrin sera (736.5, 738.5, 739.5, and 163.3) or their respective prebleeds for 1 h. For immunoprecipitation of vimentin, the precleared supernatants were incubated with a combination of antivimentin sera (Chemicon International, Inc.; Sigma Chemical Co.). Protein A agarose was added and the extracts incubated for 1 h. The beads were washed four times with buffer A (0.1% Triton-X 100, 150 mM NaCl, 10 mM Tris-HCl, pH 8.0) and directly solubilized in the appropriate buffer for one- or two-dimensional analysis.
Two-dimensional Gel Electrophoresis
Two-dimensional gel electrophoresis was carried out as described by O'Farrell et al. 1977. Isoelectric focusing was carried out in the first dimension by using ampholytes pH 3-10 (Bio-Rad Laboratories), and setting a pH gradient from 4–8. Proteins were resolved in the second dimension on a 7.5% SDS–polyacrylamide gel. The antifimbrin complex immobilized on protein A agarose beads was solubilized in SDS-IEF buffer (0.5% SDS, 9.5 M urea, 2% ampholytes pH 3-10, and 5% 2-mercaptoethanol). After 10 min, an equal aliquot of Garrel's buffer (4% NP-40, 9.5 M urea, 2% ampholytes pH 3-10, and 5% 2-mercaptoethanol) was added and incubated at 37°C for 15 min. Samples were loaded at the basic end and allowed to electrophorese for 8,000 Vh. After isoelectric focusing was complete, the IEF gels were equilibrated in SDS sample buffer, and then electrophoresed in the second dimension. The gels were finally silver stained (Shevchenko et al. 1996).
Protein Identification
The silver-stained spots were excised from the gel, washed, and processed as described by Shevchenko et al. 1996. The protein was reduced with 10 mM DTT and alkylated with 55 mM iodoacetamide. The gel piece was finally dried in a Speed-Vac and rehydrated at 4°C in digestion buffer (50 mM NH4HCO3) containing trypsin (Boehringer Mannheim). Excess trypsin was removed and the gel incubated overnight at 37°C. Digested peptides were extracted with 5% formic acid and 50% acetonitrile. The pooled extracts were dried, dissolved in 5 μl of 5% formic acid and 50% acetonitrile, mixed in a 1:1 ratio with either α-cyano-4-hydroxy-trans-cinnamic acid or 3,5 dimethoxy-4-hydroxy cinnamic acid (Aldrich Chemical Co.), and then spotted onto the target plate. All mass spectra were obtained on the Voyager Elite mass spectrometer (DE-STR; PerSeptive Biosystems) operated in the reflector mode. A list of monoisotopic peptide masses (>1,000 and <3000 Da) was imported into PepFrag and MS-Fit that searched the Swiss-Prot or GenPept databases. The following parameters were used in searches: cysteines were modified by aminocarboxymethylation, proteolytic digestion with trypsin (3–4 incompletes), mass tolerance of 1 Da, and molecular weight range of 10 kD.
In other experiments, proteolytic fragments of fimbrin and vimentin were identified by microsequencing of electroblotted fragments following previously published protocols (Matsudaira 1987). Peptides were sequenced using a microsequencer (model 477A; Applied Biosystems).
Overlay Binding Assays
The fimbrin and vimentin concentrations were obtained by measuring absorbance at 280 nm and by SDS-PAGE. Protein was mixed with Sulfo-NHS-Biotin (Pierce Chemical Co. ) at a molar ratio of 1:20 and coupled following the supplier's protocol. Unreacted biotin was removed and the derivatized protein concentrated on a 30-kD cut-off microconcentrator (Amicon). Typically, 100% of the molecules were derivatized as assayed by binding to streptavidin-agarose.
The overlay binding assay was performed essentially as described by Merdes et al. 1991. Electrophoresis of protein samples was performed in duplicate; one of the gels was stained and the other electroblotted onto polyvinylidene difluoride (PVDF) (Millipore Corp.). The blot was blocked with 1% nonfat dry milk in TBST (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.02% Tween 20) for 1 h. For the fimbrin overlay assay, the blot was incubated overnight with labeled fimbrin in 1% nonfat dry milk in TBST. For the vimentin overlay assay, the blot was incubated overnight with labeled vimentin in 1% nonfat dry milk in a low ionic strength buffer (8.03 mM NaH2PO4, 1.47 mM KH2PO4, 0.2 mM CaCl2 and 1% Triton X-100, pH 7.2). The blot was washed with TBST, incubated for 1 h with the ABC elite vector reagent (Vector Labs, Inc.), washed again with TBST, and finally developed using the ECL kit (Amersham).
In Vitro Binding with Soluble Vimentin
Purified vimentin was purchased (Cytoskeleton), whereas recombinant fimbrin and vimentin were expressed and purified as described above. Vimentin was kept in an unpolymerized state by disassembling filaments in 8 M urea, 5 mM sodium phosphate, pH 7.2, 0.2% 2-mercaptoethanol, and 1 mM PMSF. The sample was dialyzed overnight against 5 mM sodium phosphate, pH 7.4, containing 0.2% 2-mercaptoethanol and 0.2 mM PMSF. The protein concentration was adjusted to ∼1 mg/ml and incubated with precleared fimbrin (50 μg each) at 37°C for 1 h in 1 ml of low ionic strength buffer containing 8.03 mM NaH2PO4, 1.47 mM KH2PO4, 0.2 mM CaCl2 and 1% Triton X-100, pH 7.2. Antifimbrin or preimmune serum was added and the proteins were incubated for an additional 1 h. Protein A–Sepharose was added for 1 h and the beads were finally washed three times with buffer A and directly solubilized in SDS–sample buffer. The samples were resolved on SDS-PAGE, electroblotted onto a PVDF membrane, and probed with an antivimentin HRP-linked antibody (Affinity Biologicals Inc.).
Polymerization of Vimentin and Binding Assay with Fimbrin
Vimentin was diluted to a concentration of 0.5 mg/ml in a total volume of 100 μl using assembly buffer (6 mM phosphate buffer, pH 7.4, 3 mM KCl, 0.2% 2-mercaptoethanol, 0.2% PMSF). Polymerization was achieved by adding 3 μl of 5 M NaCl and incubating at 37°C for 1 h. For cosedimentation assays, preformed vimentin IFs were incubated for an additional hour with fimbrin maintained at concentrations from 0.25 to 1 mg/ml. In other experiments, vimentin and fimbrin were mixed together and IF polymerization, then initiated in the assembly buffer. The proteins were collected by ultracentrifugation at 100,000 g for 30 min and analyzed by SDS-PAGE.
Estimation of Kd
Purified vimentin (2.5 μg) was applied to nitrocellulose membranes. The blots were placed into wells of a 12-well tissue culture plate (Costar Corp.) and an overlay assay with labeled fimbrin (0.1–1 μM) was performed as described earlier. The amount of labeled protein that bound was estimated by densitometric scanning (GS-700 Densitometer; Bio-Rad Laboratories) and quantified using the molecular analyst software (Bio-Rad Laboratories). Nonspecific binding of biotinylated fimbrin was determined by measuring the amount of binding to nitrocellulose containing no vimentin. Specific binding was determined by subtracting nonspecific binding from the total binding. Scatchard analysis was carried out to estimate the K d of binding. Plot shown gives on the vertical axis bound/free fimbrin and on the horizontal axis bound fimbrin (pmols). The amount of fimbrin that bound in pmols was obtained from a standard curve of known amounts of biotinylated fimbrin directly spotted onto nitrocellulose.
Fimbrin Proteolysis
Small amounts of Lys-C (0.25 μg) were incubated with purified fimbrin (50 μg) under nondenaturing conditions in 50 μl of 25 mM Tris, pH 7.7, and 1 mM EDTA. Aliquots were removed at 1, 10, 15, and 30 min, quenched by the addition of PMSF, and then boiled in SDS–sample buffer. The digested products were resolved on SDS-PAGE gels that were either silver stained or electroblotted onto PVDF.
Immunofluorescence Microscopy
For immunofluorescence studies, cells were allowed to attach onto glass coverslips (VWR Scientific) for 1–48 h, gently rinsed in PBS, and fixed with 4% paraformaldehyde (PFA) in PBS for 10 min. After three washes with PBS, the cells were permeabilized by incubating with 0.1% Triton X-100 in PBS for 2 min before immunostaining. To prepare detergent-extracted cells, adherent cells were first incubated with ice-cold 10 mM Pipes buffer, pH 6.8, containing 0.5% Triton X-100, 300 mM sucrose, 100 mM KCl, 3 mM MgCl2, 10 mM EGTA, 2 mM PMSF, and 50 μM sodium vanadate for 3 min, washed with the same buffer, and then fixed with 4% PFA.
Nonspecific protein absorption was inhibited by incubating the cells for 1 h in PBS containing 3% BSA, 0.2% Tween 20, and 0.2% fish gelatin. Coverslips were incubated for 1 h at 37°C with affinity-purified rabbit–fimbrin antibody (737.4a) diluted 1:100 and goat–vimentin antibody (Sigma Chemical. Co.; Chemicon International, Inc.) diluted 1:50. The coverslips were washed with PBS and the cells were incubated for 1 h at 37°C with secondary antibodies, FITC-conjugated donkey anti–rabbit and Texas red–conjugated donkey anti–goat (Jackson ImmunoResearch Laboratories). After three washes with PBS, the coverslips were directly mounted in Vectashield (Vector Labs, Inc.) and examined. Cells examined for actin distribution were stained with rhodamine phalloidin (Molecular Probes Inc.).
Cells were imaged by epifluorescence microscopy using a Nikon TE300 microscope with 60 and 100× oil immersion lenses. The images were recorded with a Hammatsu Orca CCD camera and analyzed with the Open Lab software program (Improvision Inc.). For deconvolution microscopy, the cells were viewed on a Nikon Eclipse 800 fluorescence microscope with a 100× oil immersion lens. Image stacks were recorded at 134-nm intervals (z series) with a Hammatsu Orca CCD camera and analyzed with the CellScan software (Scanalytics) or Metamorph imaging system (Universal Imaging Corp.) using the exhaustive photon reassignment algorithm. The images were reconstructed in three dimension using either the acquisition or Imaris software (BitPlane). Colocalized proteins were identified by using the colocalization algorithm in Imaris, where voxels containing the two different signals were extracted. The images were finally processed with Adobe Photoshop.
Results
A Fimbrin–Vimentin Complex in Adherent Macrophages
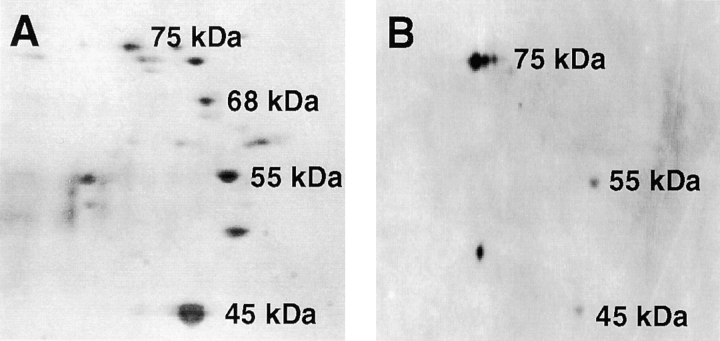
In this paper, we investigated the mechanism that targeted fimbrin to podosomes and filopodia at the cell–substratum interface of adherent macrophage cells. Because cytoskeleton–matrix interactions could involve different sets of proteins than in the cortical cytoskeleton, our initial experiments examined whether fimbrin is in a precipitable complex with other proteins in adherent macrophage cells. In Triton-soluble extracts of adherent P388D1 cells, we found four polypeptides of molecular weights, 45, 55, 68, and 75 kD that coprecipitated in immune complexes with four different fimbrin-specific antisera (Fig. 1 A). This interaction with fimbrin is specific, because exogenously added fimbrin inhibited the immunoprecipitation of the 45, 55, and 75 kD (not shown). Other polypeptides seen on the gel were also present in immunoprecipitates of the preimmune sera and, therefore, were not characterized.
Figure 1.
Identification of fimbrin-binding proteins present in the Triton X-100 extracts from adherent P388D1 cells. (A) Proteins immunoprecipitated with fimbrin antibodies were separated on two-dimensional gels and detected by silver stain. Four polypeptides of molecular mass 45,000, 55,000, 68,000, and 75,000 Da were precipitated with fimbrin antisera and were not present in control antisera. The four proteins were identified from their tryptic mass fingerprints as actin (45,000), vimentin (55,000), fimbrin (68,000), and HSP-70 (75,000). Other polypeptides on the gel were also present in the control antisera and were not characterized further. (B) In a fimbrin overlay assay of proteins electroblotted onto PVDF membranes, only the 45-, 55-, and 75-kD proteins bind the biotinylated fimbrin probe.
Having detected several potential fimbrin-binding proteins, we proceeded to identify them as actin (45 kD), vimentin (55 kD), and the heat shock protein, Hsp70 (75 kD). Identification was based on a search of sequence databases (PepFrag and MS-FIT) using the tryptic mass fingerprint of each polypeptide and confirmed by immunoblots using commercially available antisera that are specific for each protein (not shown). Finally, to confirm that fimbrin bound the proteins directly we also probed electroblots of two-dimensional gels with biotinylated fimbrin. On the blots, fimbrin bound actin, vimentin, and Hsp70 (Fig. 1 B). Binding to these proteins was inhibited by an excess of unlabeled fimbrin (not shown). Although Hsp70 appeared to bind fimbrin with a higher affinity, we studied the fimbrin–vimentin interaction because of its potential role in modulating the assembly and structure of the actin and IF cytoskeletons.
Characterization of Fimbrin–Vimentin Binding
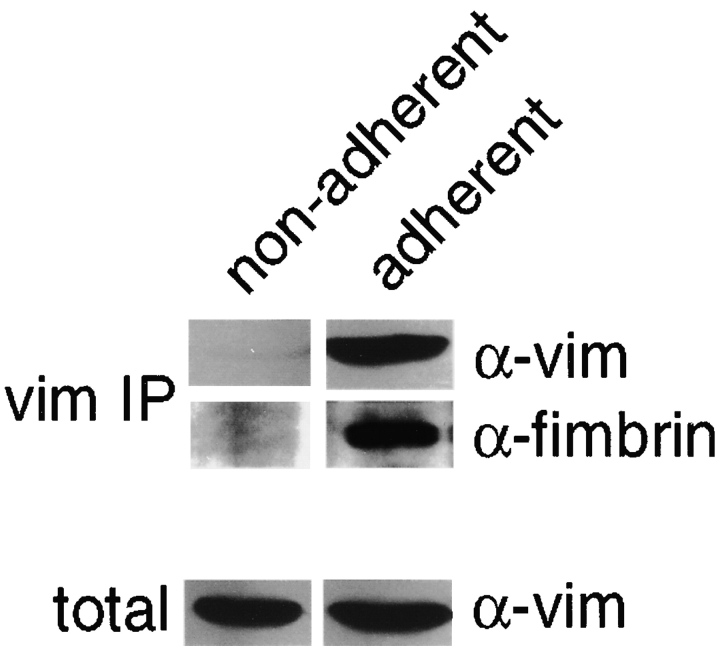
Because the fimbrin–vimentin complex is found in a Triton X-100 extract of adherent cells, we also investigated whether the complex was present in nonadherent cells. Although the total amount of vimentin in adherent and nonadherent cells is identical, we could immunoprecipitate a detergent soluble pool of vimentin only from adherent cells using vimentin-specific antibodies (Fig. 2). Furthermore, a fimbrin-specific antibody detected fimbrin only in vimentin immunoprecipitates from adherent cells. Reciprocal immunoprecipitations with fimbrin-specific antibodies confirmed the presence of an extractable complex in adherent cells but not in nonadherent cells (not shown). This extractable fraction of vimentin in adherent cells was estimated to be ∼1% of the total protein.
Figure 2.
Fimbrin–vimentin complexes in adherent P388D1 cells. Triton X-100 extracts from adherent and nonadherent cells were prepared and vimentin immunoprecipitates were probed with vimentin- or fimbrin-specific antibodies. Fimbrin coprecipitated with vimentin in adherent cells, but was not detected in nonadherent cells. The same cells were directly solubilized in sample buffer, resolved on SDS-PAGE, electroblotted, and probed to show equivalent amounts of total vimentin. The detergent extracted pool of vimentin present in adherent cells was estimated to be ∼1% of the total vimentin.
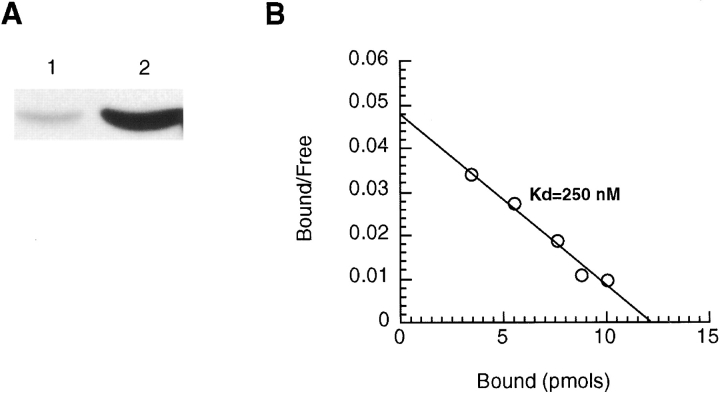
To understand the nature of the fimbrin–vimentin interaction, we first tested whether fimbrin bound preassembled vimentin filaments using a standard high speed pelleting assay. However, this assay was unable to detect fimbrin in pellets of vimentin filaments (not shown). Next, we tested the binding of fimbrin to unpolymerized vimentin subunits using an immunoprecipitation assay. From a purified mixture of fimbrin and vimentin, vimentin is detected in immunoprecipitates using a fimbrin antibody and not in the preimmune serum (Fig. 3 A). To characterize this interaction more carefully, we measured the binding of soluble fimbrin to vimentin that was immobilized on nitrocellulose membranes. On a Scatchard plot (Fig. 3 B), we estimated the K d of fimbrin binding to vimentin to be 0.25 μM. From the intercept at the x-axis, fimbrin binds 3.8 molecules of vimentin. A simple interpretation of the data is that fimbrin binds a tetramer of vimentin.
Figure 3.
Fimbrin binding to unpolymerized vimentin. (A) Under conditions that prevent vimentin polymerization, aliquots from mixtures of pure recombinant vimentin and fimbrin (50 μg each) were incubated with preimmune serum (lane 1) or antifimbrin (lane 2) antibodies. When the immunoprecipitates were probed with an antivimentin antibody, only the fimbrin immunoprecipitate contained vimentin. (B) The K d of the fimbrin–vimentin interaction was obtained by dot blot assay and converting the data into a Scatchard plot. The K d was calculated to be 0.25 μM and the intercept is at 1:3.8 mole ratio.
Mapping the Fimbrin- and Vimentin-binding Regions
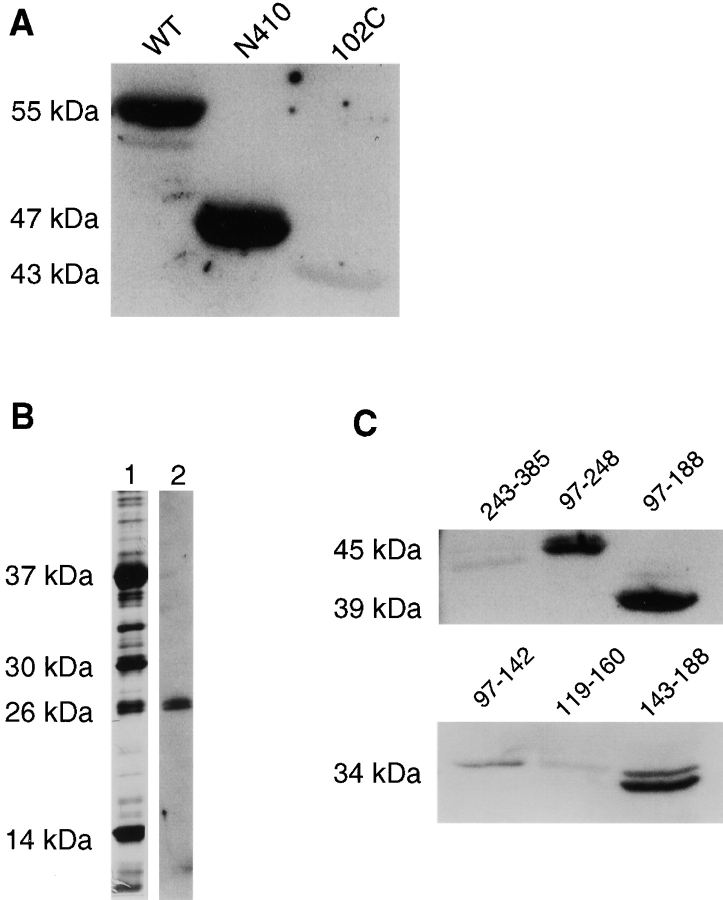
Because the binding of fimbrin to unpolymerized vimentin is a novel finding, we proceeded to investigate whether the interaction can be localized to discrete domains in both proteins using proteolytic fragments as well as different deletion constructs. In an overlay assay (Fig. 4 A), biotinylated fimbrin is shown to bind to full-length unpolymerized vimentin (WT) and vimentin deleted of its COOH terminus (N410), but does not bind to vimentin deleted of its NH2 terminus (102C). Similar results were obtained with an overlay assay of the NH2- and COOH-terminal fragments of vimentin that were generated by 2-nitro-5-thiocyanobenzoate cleavage at the single cysteine residue present in the molecule (not shown). Binding between the two proteins was not affected by the ionic strength of the assay and comparable levels of binding were detected in either low (5 mM sodium phosphate) or high (150 mM NaCl) ionic strength solutions. These results suggest that fimbrin binds specifically to the NH2-terminal region of vimentin.
Figure 4.
CH1 domain of fimbrin binds the NH2 terminus of nonassembled vimentin. (A) In vitro expressed and purified wild type (WT), COOH-terminally deleted (N410) or NH2-terminally deleted (102C) vimentin were resolved on SDS-PAGE, transferred to PVDF, and probed with labeled fimbrin. Fimbrin binds wild type (WT) and the COOH-terminally deleted vimentin (N410) but not the NH2-terminal deleted vimentin (102C). (B) Limited proteolysis of fimbrin with Lys-C and the fragments were detected with silver stain (lane 1), electroblotted, and probed with biotinylated vimentin (lane 2). Vimentin bound a 26-kD fragment. (C) Overlays of fimbrin deletion constructs using biotinylated vimentin. Vimentin bound bands that contained residues 143–188 in the CH1 domain.
To map the vimentin-binding region on fimbrin, we conducted overlay assays on fimbrin proteolyzed with lys-C (Fig. 4 B). In these experiments, a low ionic strength buffer was used to keep vimentin from polymerizing. Biotinylated vimentin is seen to specifically bind to a 26-kD domain of fimbrin. The 26-kD fragment was sequenced and shown to be part of ABD1. To narrow the vimentin binding site on actin binding domain 1, we tested binding of biotinylated vimentin to various truncations of the ABD1 domain (Fig. 4 C). Using the overlay assay, we found that vimentin binds any part of the CH1 domain that contained residues 143–188 (Table ).
Table 1.
Summary of Vimentin Binding to Fimbrin Polypeptides
| Sequence | Region of fimbrin | Binding |
|---|---|---|
| Fimbrin |
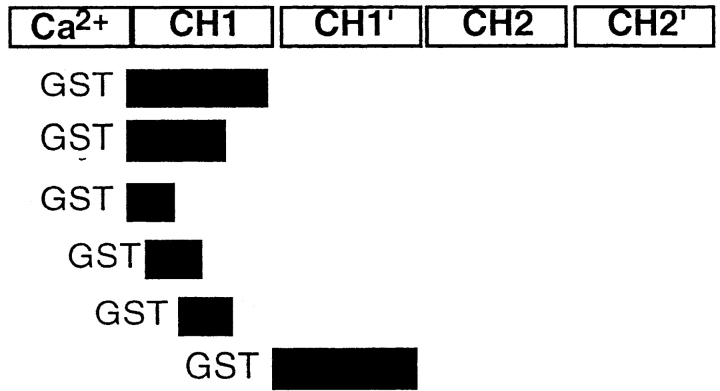
|
+ |
| 97–248 | + | |
| 97–188 | + | |
| 97–142 | − | |
| 119–160 | − | |
| 143–188 | + | |
| 243–385 | − |
Immunolocalization of Fimbrin and Vimentin in IC-21 and P388D1 Cells
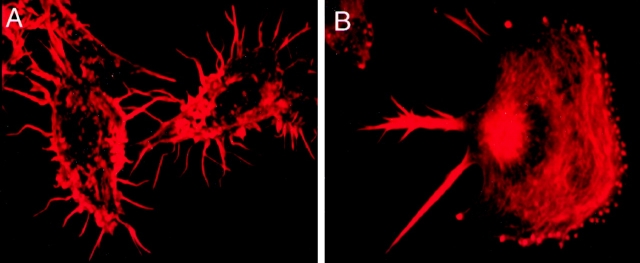
Because the biochemical experiments strongly suggested that fimbrin is in a complex with vimentin subunits, we carried out a series of immunocytological studies to identify how the proteins were distributed and to determine where this complex is localized. If fimbrin and vimentin are in a complex, they should colocalize to the same structures and compartments in the cell. Using fluorescence microscopy, we first examined the distribution of filamentous actin (F-actin) as well as fimbrin and vimentin in two macrophage cell lines; highly differentiated, motile IC-21 cells and moderately differentiated, nonmotile P388D1 cells. In nonmotile P388D1 cells, phalloidin staining was localized in filopodia and cell surface microvilli and the cell body (Fig. 5 A). However, P388D1 cells do not organize the actin cytoskeleton into podosomes, and the dotted pattern characteristic of podosomes was absent in these cells. On plated IC-21 cells (Fig. 5 B), F-actin is detected in clusters of small dots, podosomes, or rosette adhesions at the leading edge of the cell, in the cell body, and in retraction fibers at the trailing end of the cell. This staining pattern is similar to that observed when primary mouse macrophages are stained for actin (Messier et al. 1993). Thus, the actin cytoskeleton is organized differently in the two cell lines.
Figure 5.
F-actin distribution in the two macrophage cell lines by epifluorescence microscopy. (A) Nonmotile P388D1 cells stained with rhodamine phalloidin show F-actin staining in filopodia, cell surface microvilli and the cell body. (B) Staining of motile IC-21 cells with rhodamine phalloidin detected F-actin in clusters of small adhesions at the leading edge of the cell, in the cell body, and in retraction fibers at the trailing end of the cell.
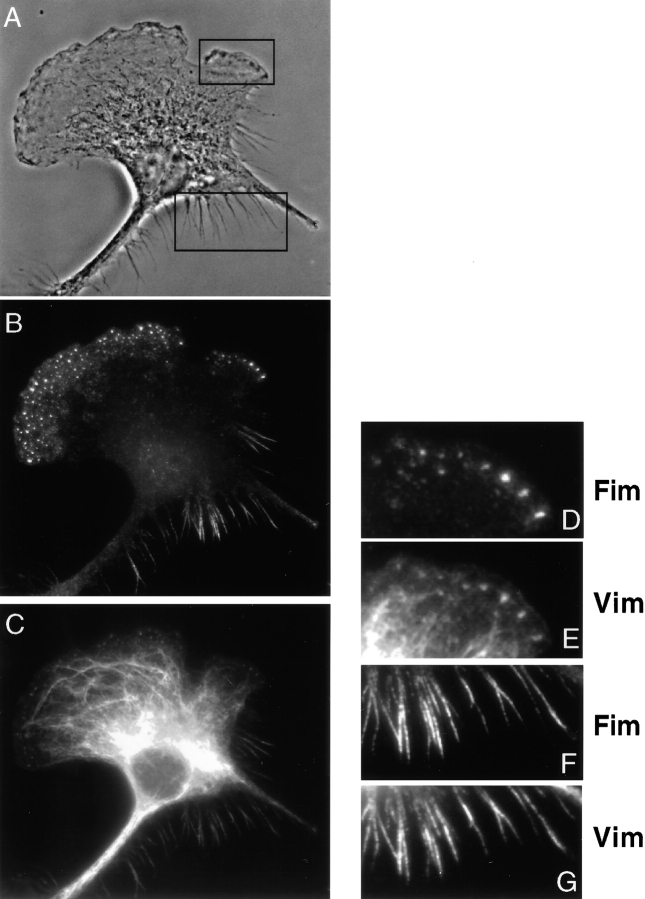
We examined the distribution of fimbrin and vimentin in the two macrophage cell lines by fluorescence deconvolution microscopy. Double staining of P388D1 cells revealed fimbrin in filopodia and in the perinuclear region of the cell (Fig. 6 A), whereas vimentin localized to filopodia and the cell body (Fig. 6 B). To obtain colocalized signals we imported image stacks for each fluorescent pattern into an image processing software, Imaris, and voxels (134 nm) that contained the two signals were extracted and displayed in yellow. Fimbrin and vimentin are seen to colocalize in the filopodia (Fig. 6 C). In IC-21 cells, fimbrin is found in podosomes, retraction fibers, and in the perinuclear region of the cell (Fig. 6 D), whereas vimentin stained podosomes, retraction fibers, and the cell body (Fig. 6 E). The two image stacks were also processed in Imaris, and voxels containing the two fluorescent signals (displayed in yellow) show that the proteins colocalized in podosomes (arrow), retraction fibers, and in the perinuclear region (Fig. 6 F). The colocalization of both proteins on the basal surface was confirmed from side views of a three-dimensional reconstruction of the cell (not shown). These results show that fimbrin and vimentin colocalize to actin-rich structures, filopodia, podosomes, and retraction fibers in two different macrophage cells.
Figure 6.
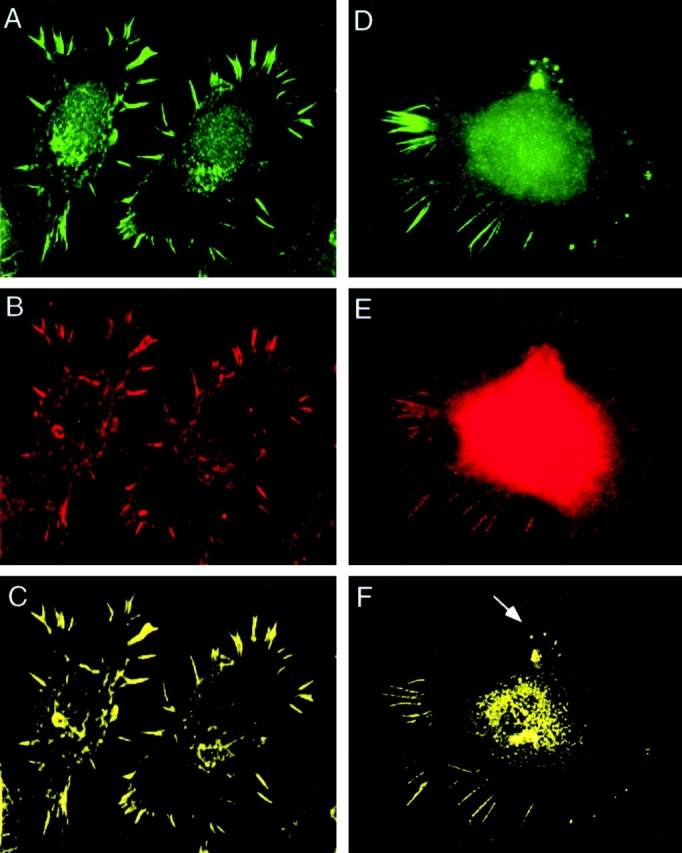
Fimbrin–vimentin localization by deconvolution microscopy. In P388D1 cells (A–C), immunostaining with a fimbrin antibody (A) labeled the filopodia and the perinuclear region. Immunostaining for vimentin (B) in the same cells localized the protein in filopodia and in the cell body. Image stacks for each of the signals were imported into an image processing software, Imaris, and reconstructed in three dimension. The colocalizing module in Imaris was used to extract voxels (134 nm) containing the two signals that are displayed in yellow. The two proteins are seen to colocalize in filopodia, with some perinuclear staining (C). In IC-21 cells (D–F), podosomes, perinuclear region, and retraction fibers at the trailing end of the cell were labeled with fimbrin antibodies (D). Immunostaining of vimentin in the same cells (E) localized the protein in the cell body and in retraction fibers. Using Imaris, the two proteins are shown to colocalize in retraction fibers and in podosomes (arrow) with some costaining in the perinuclear region (F).
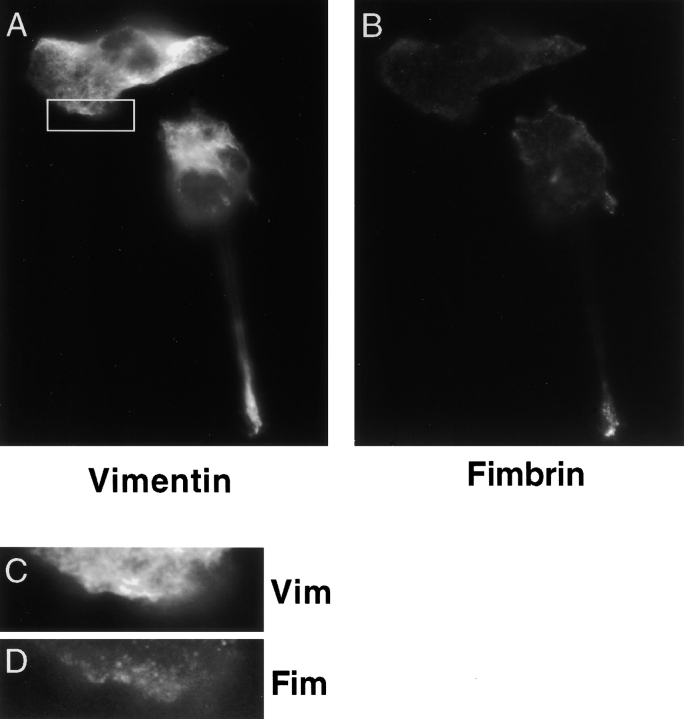
Our biochemical studies detected fimbrin and vimentin in the Triton X-100 extract of IC-21 and P388D1 cells. To locate those structures that contain this extractable complex, we also examined the distribution of fimbrin and vimentin after Triton X-100 treatment. Before extraction, the complex was examined on freshly plated cells. Macrophages that had just attached and were beginning to spread show vimentin and fimbrin staining primarily in the perinuclear region (not shown). Half an hour after plating, the two proteins were found in numerous common foci in regions that were actively spreading (Fig. 7, A–E). However, the majority of the vimentin network is in the perinuclear region. 3 h after plating, the cells were well polarized and the complex was now restricted to foci in that region of the cell that was actively expanding and to retraction fibers at the trailing end of the cell (Fig. 8, A–G). After Triton X-100 extraction, IC-21 cells show complete loss of staining of fimbrin and vimentin from both retraction fibers and podosomes (Fig. 9, A–D). Similarly, in P388D1 cells, both proteins were less abundant or undetected in filopodia but were present in the cell body (not shown).
Figure 7.
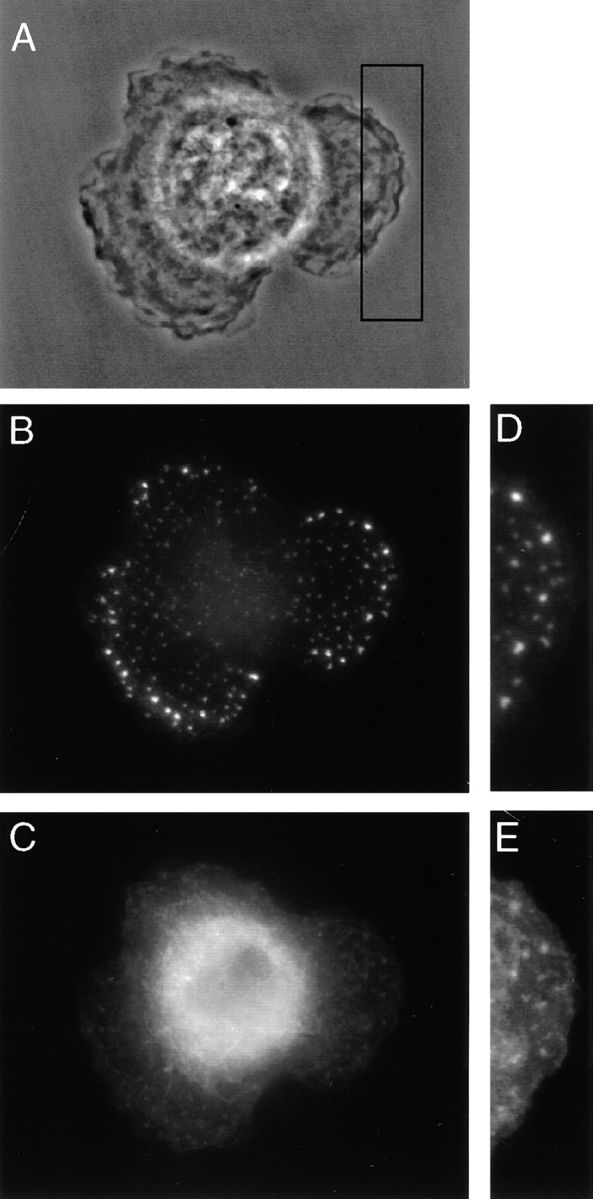
Localization of the fimbrin–vimentin complex in spreading cells by phase and epifluorescence microscopy. Half hour after plating, IC-21 cells are seen spreading out in all directions (A). Fimbrin (B and D) and vimentin (C and E) are detected in common foci in regions of the cell that are actively spreading. The vimentin network is collapsed around the nucleus.
Figure 8.
Localization of the fimbrin–vimentin complex in polarized IC-21 cells. 3 h after attachment, the cells show, by phase microscopy, an asymmetrical morphology (A). Using epifluorescence microscopy the fimbrin–vimentin complex is shown to be restricted to foci present at the cells leading edge where the lamella is actively expanding (D and E). The vimentin network is extended out in the direction of the leading edge (C). The fimbrin–vimentin complex is also detected in retraction fibers at the trailing end of the cell (F and G).
Figure 9.
IC-21 cells are extracted with Triton X-100 before fixation, and then stained for fimbrin (B and D) and vimentin protein (A and C). Cells extracted with detergent show complete loss of staining for both proteins in podosomes and retraction fibers supporting the detergent extractable nature of the complex in these two structures. However, the vimentin network is present in the cell body after extraction (A).
Discussion
Our studies describe an adhesion-dependent relationship between the actin and intermediate filament cytoskeletons. We present four independent observations that document a specific interaction between an actin cross-linking protein, fimbrin, and an IF protein, vimentin. First, the two proteins coprecipitate from Triton-X 100–extracted cells with either fimbrin- or vimentin-specific antibodies. Second, binding assays with recombinant proteins restrict the binding region between residues 143–188 of fimbrin and the head domain of vimentin. Third, the two proteins colocalize in podosomes, filopodia, and retraction fibers. Finally, both proteins are extracted from these structures by the same detergent treatment. These biochemical and morphological observations are strong evidence of a fimbrin–vimentin complex that is found in ventral structures where a macrophage adheres to the substratum.
The association between fimbrin and vimentin, reported in this study, is a new finding. Based on previous studies, fimbrin was shown to bind and cross-link actin filaments. This function was consistent with its localization in actin bundles that support cell surface microvilli (Bretscher and Weber 1980; Matsudaira 1991). However, early studies also detected fimbrin at the cell–matrix interface (Bretscher and Weber 1980; Marchisio et al. 1987), which suggested that fimbrin had a function associated with cell adhesion. This suggestion was reinforced by our finding that during embryogenesis L-fimbrin is transiently expressed and localized to the basal surface of embryonic enterocytes, whereas T- and I-fimbrin and the majority of F-actin are in the developing brush border at the apical surface of the same cells (Chafel et al. 1995). Because IFs are now recognized components of focal adhesions (Green et al. 1986; Bershadsky et al. 1987; Geiger et al. 1987; Seifert et al. 1992), interactions with fimbrin could explain how vimentin is differentially targeted to cell adhesion structures where it can assemble into IFs. In addition, association with vimentin that may be transported to adhesion sites in a microtubule-dependent mechanism (Prahlad et al. 1998; Yoon et al. 1998) can explain how fimbrin is targeted to adhesion sites where it can cross-link actin filaments into a bundle.
The fimbrin–vimentin interaction is unusual because fimbrin binds a vimentin subunit and not a filament. Typically, cytoskeletal networks are cross-linked by proteins that bind polymerized filaments (Svitkina et al. 1996; Yang et al. 1996). For example, elegant immuno EM studies show plectin cross-bridges between assembled IFs and MTs (Svitkina et al. 1996). Plectin is also implicated in binding to actin filaments through an actin-binding domain located at the NH2 terminus, whereas the IF-binding domain is found at the COOH terminus (Wiche et al. 1993; McLean et al. 1996). However, this is not the case for fimbrin. Its interaction with a subunit of vimentin is suggested by the following: the 1:4 stoichiometry of binding (Fig. 3 B), the absence of substantial fimbrin colocalization with vimentin filaments in the cell body, and the inability of fimbrin to bind polymerized vimentin in pelleting assays (Correia, I., and P. Matsudaira, unpublished data). The detergent-extracted fraction most likely includes the pool of unassembled vimentin subunits in the cell (Soellner et al. 1985) that are believed to be tetramers of vimentin. The identity of the vimentin subunit as a binding partner of fimbrin is also consistent with the low percentage (1%) of this species in the supernatant (Fig. 2). Thus, the interaction with a vimentin subunit and not a filament suggests that the complex has a function other than to cross-link the actin and IF cytoskeletons.
Binding to vimentin subunits places fimbrin in a unique position to influence the assembly and organization of both the actin and IF cytoskeletons. We speculate that the fimbrin–vimentin complex plays an early role in the assembly of the actin and vimentin cytoskeleton in filopodia and podosomes. This speculation is supported by our biochemical observation that the fimbrin–vimentin interaction in P388D1 cells is adhesion-dependent. The time course of the localization of fimbrin and vimentin in these structures, obtained by epifluorescence microscopy, also suggests that the complex is a transient structure that is involved early in cell adhesion. During the early stages of attachment of motile IC-21 cells, we detected costaining of fimbrin and vimentin in podosomes of expanding lamella. After 3 h, fimbrin does not become associated with the vimentin network of filaments (Fig. 7 and Fig. 8). Because fimbrin does not bind assembled vimentin filaments in vitro other proteins may assist in anchoring the vimentin network at mature adhesion sites. These observations are relevant as studies obtained from time-lapse and interference reflection microscopy indicate a preformed complex of cytoskeletal proteins that nucleate the recruitment of adhesive molecules before anchoring the lamellipodia and filopodia to the substratum (De Pasquale and Izzard 1987; Izzard 1988). Although the compositions of the nucleating proteins were not described, the fimbrin–vimentin complex is in the right place and at the right time to play this role.
This model of fimbrin–vimentin function is compatible with earlier reports from Tao and colleagues (Mabuchi et al. 1997), who described a similar interaction between desmin tetramers and calponin in smooth muscle cells. They proposed the idea that an IF subunit and an actin-binding protein may be involved in the assembly of IFs at dense bodies. We agree with their interpretation. In fact, it is alluring to consider that the interactions involve cell adhesions. As discussed in the following section, an interaction between an IF protein, desmin, or vimentin, and a protein with a CH domain, calponin, or fimbrin may represent a highly conserved function that may be related to the establishment of cell adhesion structures.
The fimbrin–vimentin complex is biochemically distinguished from the rest of the actin and IF cytoskeletons by its extraction properties. The critical observation is that fimbrin and vimentin are extracted from a subset of structures (podosomes, filopodia, and retraction fibers) by detergent. Biochemical quantitation shows the detergent-extracted population represents ∼1% of the total vimentin in the cell. Most vimentin is in a detergent-insoluble fraction, and fluorescence microscopy localizes the insoluble vimentin to filaments in the cell body and around the nucleus. Thus, the vimentin–fimbrin complex found in early cell adhesion structures is biochemically and structurally distinct from the vimentin network in the cell body. This distinction may provide useful approaches for studying the function of the complex in live cells.
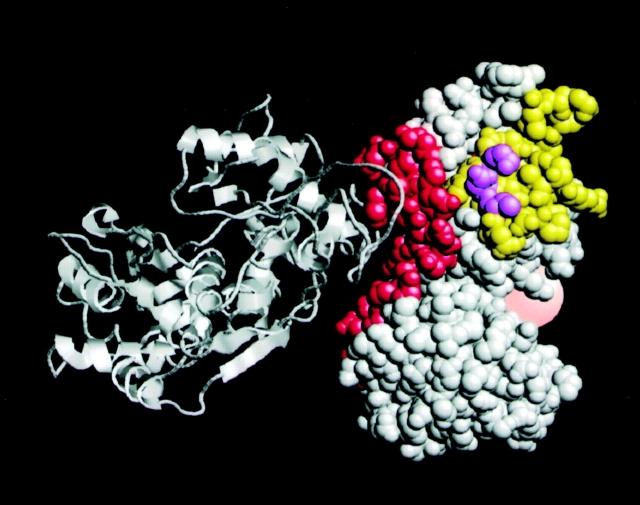
The biochemical and structural properties of the fimbrin–vimentin complex may be explained by the location of fimbrin and vimentin binding sites on the proteins. Our overlay binding assays now tentatively map the region of fimbrin–vimentin interaction to residues 143–188 of the NH2-terminal actin-binding domain of fimbrin and the head domain of vimentin. These sites lie within interesting regions of both proteins. Fimbrin binds a pair of actin filaments using actin-binding domains that are located in the NH2- and the COOH-terminal regions of the molecule (Hanein et al. 1997). Quantitative modeling of a fimbrin x-ray structure to an EM reconstruction of fimbrin bound to actin filaments predicts that actin binds fimbrin through interactions at the two CH domains in the NH2-terminal actin-binding domain, ABD1 (Hanein et al. 1998). Vimentin also binds fimbrin on ABD1, opposite to the actin-binding site, but on the same side where the regulatory calcium-binding domain (residues 1–100) and the COOH-terminal actin-binding domain (ABD2) are located (Fig. 10). This location poses interesting implications for actin bundling. Although we have not yet studied the ability of the fimbrin–vimentin complex to bind and cross-link actin filaments, we suspect that vimentin binding to fimbrin may interfere with the ability of fimbrin to cross-link actin.
Figure 10.
Distinct binding sites of actin and vimentin on the CH1 domain of fimbrin. A ribbon model of actin (white) is shown docked on the space filled model of ABD1 of fimbrin (white). The putative vimentin binding site (lime) is distinct from the actin-binding site (red) and the proposed model easily accommodates the NH2-terminal calcium–binding domain (transparent red) and ABD2 of fimbrin (not shown) in the presence of vimentin. Loops that overlap in the two binding sites are depicted in purple.
The fimbrin binding site lies on an important domain of vimentin. The NH2-terminal domain is absolutely essential for incorporation of vimentin subunits into protofilaments and higher order filaments, and deletion of this domain prevents assembly of vimentin filaments (Traub and Vorgias 1983; Perides et al. 1987; Shoeman et al. 1990; Herrmann et al. 1996). Furthermore, phosphorylation of the IF network on the NH2- and COOH-terminal domains (head and tail domains) is a well established means for the disassembly of the IF network (Inagaki et al. 1996). Interestingly, preliminary studies carried out in vivo indicate that the fimbrin–vimentin complex is phosphorylated on vimentin and not on fimbrin (not shown). Although we have not localized the sites of phosphorylation, fimbrin bound to the NH2-terminal domain of vimentin may play a role in regulating vimentin assembly by either directly preventing incorporation of its subunits into filaments, or indirectly, by modulating the addition or removal of phosphate groups.
Acknowledgments
I. Correia and P. Matsudaira thank Dorit Hanein and Niels Volkmann (both from Burnham Institute) for their help in generating 10, and Itay Rousso (Whitehead Institute) for help with the Imaris software (BitPlane).
This work was supported by National Institutes of Health grants to P. Matsudaira and R. Goldman and was conducted at the W.M. Keck Foundation Microscopy Facility at the Whitehead Institute.
Footnotes
1.used in this paper: ABD, actin-binding domain; CH, calponin homology; F-actin, filamentous actin; GST, glutathione-S-transferase; IF, intermediate filament; MF, microfilament; MT, microtubule; PVDF, polyvinylidene difluoride
References
- Bershadsky A.D., Tint I.S., Svitkina T.M. Association of intermediate filaments with vinculin-containing adhesion plaques of fibroblasts. Cell Motil. Cytoskelet. 1987;8:274–283. doi: 10.1002/cm.970080308. [DOI] [PubMed] [Google Scholar]
- Bershadsky A.D., Vasiliev J.M. Cytoskeleton 1988. Plenum Publishing Corp; New York: pp. 298 pp [Google Scholar]
- Blose S.H., Chacko S. Rings of intermediate (100 A) filament bundles in the perinuclear region of vascular endothelial cells. Their mobilization by colcemid and mitosis. J. Cell Biol. 1976;70:459–466. doi: 10.1083/jcb.70.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A., Weber K. Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J. Cell Biol. 1980;86:335–340. doi: 10.1083/jcb.86.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill S., Li S., Lyman C.W., Church D.M., Wasmuth J.J., Weissbach L., Bernards A., Snijders A.J. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J., Saraste M. Does Vav bind to F-actin through a CH domain? FEBS (Fed. Eur. Biochem. Soc.) Lett. 1995;374:149–151. doi: 10.1016/0014-5793(95)01098-y. [DOI] [PubMed] [Google Scholar]
- Chafel M.M., Shen W., Matsudaira P. Sequential expression and differential localization of I-, L-, and T-fimbrin during differentiation of the mouse intestine and yolk sac. Dev. Dyn. 1995;203:141–151. doi: 10.1002/aja.1002030203. [DOI] [PubMed] [Google Scholar]
- Chou Y.H., Skalli O., Goldman R.D. Intermediate filaments and cytoplasmic networkingnew connections and more functions. Curr. Opin. Cell Biol. 1997;9:49–53. doi: 10.1016/s0955-0674(97)80151-2. [DOI] [PubMed] [Google Scholar]
- Croop J., Holtzer H. Response of myogenic and fibrogenic cells to cytochalasin B and to colcemid. I. Light microscope observations. J. Cell Biol. 1975;65:271–285. doi: 10.1083/jcb.65.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Arruda M.V., Watson S., Lin C.S., Leavitt J., Matsudaira P. Fimbrin is a homologue of the cytoplasmic phosphoprotein plastin and has domains homologous with calmodulin and actin gelation proteins. J. Cell Biol. 1990;111:1069–1179. doi: 10.1083/jcb.111.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pasquale J.A., Izzard C.S. Evidence for an actin-containing cytoplasmic precursor of the focal contact and the timing of incorporation of vinculin at the focal contact. J. Cell Biol. 1987;105:2803–2809. doi: 10.1083/jcb.105.6.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil R.R. Structure and evolution of the actin crosslinking proteins. Bioessays. 1991;13:219–226. doi: 10.1002/bies.950130504. [DOI] [PubMed] [Google Scholar]
- Geiger B., Volk T., Volberg T., Bendori R. Molecular interactions in adherens-type contacts. J. Cell Sci. Suppl. 1987;8:251–272. doi: 10.1242/jcs.1987.supplement_8.14. [DOI] [PubMed] [Google Scholar]
- Goldman R.D., Knipe D.M. Functions of cytoplasmic fibers in nonmuscle cell motility. Cold Spring Harbor Symp. Quant. Biol. 1972;37:523–534. [Google Scholar]
- Goldsmith S.C., Pokala N., Shen W., Fedorov A.A., Matsudaira P., Almo S.C. The structure of an actin-crosslinking domain from human fimbrin. Nat. Struct. Biol. 1997;4:708–712. doi: 10.1038/nsb0997-708. [DOI] [PubMed] [Google Scholar]
- Green K.J., Talian J.C., Goldman R.D. Relationship between intermediate filaments and microfilaments in cultured fibroblastsevidence for common foci during cell spreading. Cell Motil. Cytoskelet. 1986;6:406–418. doi: 10.1002/cm.970060406. [DOI] [PubMed] [Google Scholar]
- Hanein D., Matsudaira P., De Rosier D.J. Evidence for a conformational change in actin induced by fimbrin (N375) binding. J. Cell Biol. 1997;139:387–396. doi: 10.1083/jcb.139.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D., Volkmann N., Goldsmith S., Michon A.-M., Lehman W., Craig R., DeRosier D., Almo S., Matsudaira P. An atomic model of fimbrin binding to F-actin and its implications for filament crosslinking and regulation. Nat Struct. Biol. 1998;5:23–28. doi: 10.1038/1828. [DOI] [PubMed] [Google Scholar]
- Hartwig J.H., Kwiatkowski D.J. Actin-binding proteins. Curr. Opin. Cell Biol. 1991;3:87–97. doi: 10.1016/0955-0674(91)90170-4. [DOI] [PubMed] [Google Scholar]
- Herrmann H., Haner M., Brettel M., Muller S.A., Goldie K.N., Fedtke B., Lustig A., Franke W.W., Aebi U. Structure and assembly properties of the intermediate filament protein vimentinthe role of its head, rod and tail domains. J. Mol. Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- Inagaki M., Matsuoka Y., Tsujimura K., Ando S., Tokui T., Takahashi T., Inagaki N. Dynamic property of intermediate filamentsregulation by phosphorylation. Bioessays. 1996;18:481–487. [Google Scholar]
- Izzard C.S. A precursor of the focal contact in cultured fibroblasts. Cell Motil. Cytoskelet. 1988;10:137–142. doi: 10.1002/cm.970100118. [DOI] [PubMed] [Google Scholar]
- Lin C.S., Park T., Chen Z.P., Leavitt J. Human plastin genes. Comparative gene structure, chromosome location, and differential expression in normal and neoplastic cells. J. Biol. Chem. 1993;268:2781–2792. [PubMed] [Google Scholar]
- Lin C.S., Shen W., Chen Z.P., Tu Y.H., Matsudaira P. Identification of I-plastin, a human fimbrin isoform expressed in intestine and kidney. Mol. Cell Biol. 1994;14:2457–2467. doi: 10.1128/mcb.14.4.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.G., Maercker C., Castanon M.J., Hauptmann R., Wiche G. Human plectinorganization of the gene, sequence analysis, and chromosome localization (8q24) Proc. Natl. Acad. Sci. USA. 1996;93:4278–4283. doi: 10.1073/pnas.93.9.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi K., Li B., Ip W., Tao T. Association of calponin with desmin intermediate filaments. J. Biol. Chem. 1997;272:22662–22666. doi: 10.1074/jbc.272.36.22662. [DOI] [PubMed] [Google Scholar]
- Marchisio P.C., Cirillo D., Teti A., Zambonin-Zallone A., Tarone G. Rous sarcoma virus-transformed fibroblasts and cells of monocytic origin display a peculiar dot-like organization of cytoskeletal proteins involved in microfilament-membrane interactions. Exp. Cell Res. 1987;169:202–214. doi: 10.1016/0014-4827(87)90238-2. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- Matsudaira P. Modular organization of actin crosslinking proteins. Trends Biochem. Sci. 1991;16:87–92. doi: 10.1016/0968-0004(91)90039-x. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Actin crosslinking proteins at the leading edge. Semin. Cell Biol. 1994;5:165–174. doi: 10.1006/scel.1994.1021. [DOI] [PubMed] [Google Scholar]
- Matsudaira P.T., Burgess D.R. Identification and organization of the components in the isolated microvillus cytoskeleton. J. Cell Biol. 1979;83:667–673. doi: 10.1083/jcb.83.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean W.H., Pulkkinen L., Smith F.J., Rugg E.L., Lane E.B., Bullrich F., Burgeson R.E., Amano S., Hudson D.L., Owaribe K. Loss of plectin causes epidermolysis bullosa with muscular dystrophycDNA cloning and genomic organization. Genes Dev. 1996;10:1724–1735. doi: 10.1101/gad.10.14.1724. [DOI] [PubMed] [Google Scholar]
- Merdes A., Brunkener M., Horstmann H., Georgatos S.D. Filensina new vimentin-binding, polymerization-competent, and membrane-associated protein of the lens fiber cell. J. Cell Biol. 1991;115:397–410. doi: 10.1083/jcb.115.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier J.M., Shaw L.M., Chafel M., Matsudaira P., Mercurio A.M. Fimbrin localized to an insoluble cytoskeletal fraction is constitutively phosphorylated on its headpiece domain in adherent macrophages. Cell Motil. Cytoskelet. 1993;25:223–233. doi: 10.1002/cm.970250303. [DOI] [PubMed] [Google Scholar]
- O'Farrell P.Z., Goodman H.M., O'Farrell P.H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Otto J.J. Actin-bundling proteins. Curr. Opin. Cell Biol. 1994;6:105–109. doi: 10.1016/0955-0674(94)90123-6. [DOI] [PubMed] [Google Scholar]
- Perides G., Kuhn S., Scherbarth A., Traub P. Probing of the structural stability of vimentin and desmin-type intermediate filaments with Ca2+-activated proteinase, thrombin and lysine-specific endoproteinase Lys-C. Eur. J. Cell Biol. 1987;43:450–458. [PubMed] [Google Scholar]
- Prahlad V., Yoon M., Moir R.D., Vale R.D., Goldman R.D. Rapid movements of vimentin on microtubule trackskinesin-dependent assembly of intermediate filament networks. J. Cell Biol. 1998;143:159–170. doi: 10.1083/jcb.143.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M. The CytoskeletonAn Introductory Survey 1986. Springer-Verlag Inc; Vienna: pp. 326 pp [Google Scholar]
- Seifert G.J., Lawson D., Wiche G. Immunolocalization of the intermediate filament-associated protein plectin at focal contacts and actin stress fibers. Eur. J. Cell Biol. 1992;59:138–147. [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–885. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shoeman R.L., Mothes E., Kesselmeier C., Traub P. Intermediate filament assembly and stability in vitroeffect and implications of the removal of head and tail domains of vimentin by the human immunodeficiency virus type 1 protease. Cell Biol. Int. Rep. 1990;14:583–594. doi: 10.1016/0309-1651(90)90038-z. [DOI] [PubMed] [Google Scholar]
- Soellner P., Quinlan R.A., Franke W.W. Identification of a distinct soluble subunit of an intermediate filament proteintetrameric vimentin from living cells. Proc. Natl. Acad. Sci. USA. 1985;82:7929–7933. doi: 10.1073/pnas.82.23.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T.M., Verkhovsky A.B., Borisy G.G. Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J. Cell Biol. 1996;135:991–1007. doi: 10.1083/jcb.135.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub P., Vorgias C.E. Involvement of the N-terminal polypeptide of vimentin in the formation of intermediate filaments. J. Cell Sci. 1983;63:43–67. doi: 10.1242/jcs.63.1.43. [DOI] [PubMed] [Google Scholar]
- Wang E., Choppin P.W. Effect of vanadate on intracellular distribution and function of 10-nm filaments. Proc. Natl. Acad. Sci. USA. 1981;78:2363–2367. doi: 10.1073/pnas.78.4.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiche G., Becker B., Luber K., Weitzer G., Castanon M.J., Hauptmann R., Stratowa C., Stewart M. Cloning and sequencing of rat plectin indicates a 466-kD polypeptide chain with a three-domain structure based on a central α-helical coiled coil. J. Cell Biol. 1991;114:83–99. doi: 10.1083/jcb.114.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiche G., Gromov D., Donovan A., Castanon M.J., Fuchs E. Expression of plectin mutant cDNA in cultured cells indicates a role of COOH-terminal domain in intermediate filament association. J. Cell Biol. 1993;121:607–619. doi: 10.1083/jcb.121.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Dowling J., Yu Q.C., Kouklis P., Cleveland D.W., Fuchs E. An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell. 1996;86:655–665. doi: 10.1016/s0092-8674(00)80138-5. [DOI] [PubMed] [Google Scholar]
- Yoon M., Moir R.D., Prahlad V., Goldman R.D. Motile properties of vimentin intermediate filament networks in living cells. J. Cell Biol. 1998;143:147–157. doi: 10.1083/jcb.143.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]