Abstract
The fission yeast Schizosaccharomyces pombe divides by medial fission through the use of an actomyosin contractile ring. Precisely at the end of anaphase, the ring begins to constrict and the septum forms. Proper coordination of cell division with mitosis is crucial to ensure proper segregation of chromosomes to daughter cells. The Sid2p kinase is one of several proteins that function as part of a novel signaling pathway required for initiation of medial ring constriction and septation. Here, we show that Sid2p is a component of the spindle pole body at all stages of the cell cycle and localizes transiently to the cell division site during medial ring constriction and septation. A medial ring and an intact microtubule cytoskeleton are required for the localization of Sid2p to the division site. We have established an in vitro assay for measuring Sid2p kinase activity, and found that Sid2p kinase activity peaks during medial ring constriction and septation. Both Sid2p localization to the division site and activity depend on the function of all of the other septation initiation genes: cdc7, cdc11, cdc14, sid1, spg1, and sid4. Thus, Sid2p, a component of the spindle pole body, by virtue of its transient localization to the division site, appears to determine the timing of ring constriction and septum delivery in response to activating signals from other Sid gene products.
Keywords: sid2, cdc7, spg1, cytokinesis, Schizosaccharomyces pombe
Cytokinesis is the cell cycle event that divides one cell into two daughter cells. It begins with the formation of the cleavage furrow in a plane perpendicular to the mitotic spindle apparatus. The cleavage furrow is rich in actin, myosin, and numerous other proteins (for review see Satterwhite and Pollard 1992). Cytokinesis occurs at the end of mitosis, after all the cellular constituents have duplicated and segregated into positions that will ensure that each daughter cell will receive one genome and any requisite machinery to start the next cell cycle. Cytokinesis must be highly regulated temporally with respect to other mitotic events so that the cleavage furrow does not cut through an undivided nucleus or incompletely separated set of chromosomes. Although quite a number of elegant studies involving micromanipulation of cells have shown that signals from either overlapping astral microtubules (Rappaport 1986) or the spindle midzone may cause initiation of cell cleavage at the proximal cortex (Wheatley and Wang 1996), identification of signaling molecules that direct furrow formation and cause cleavage to initiate has proven difficult, and will likely be facilitated by the use of genetic systems.
Schizosaccharomyces pombe is particularly well-suited for the study of cytokinesis, since these cylindrical cells divide by medial fission, using an actin- and myosin-rich structure termed the medial ring, which is analogous to the cleavage furrow in animal cells. S. pombe also provides several advantages as a model system, including an ease of genetic manipulations, that the genome sequence is nearing completion, and that previous studies have yielded a well-characterized cell cycle as well as several classes of cytokinesis mutants (for reviews see Chang and Nurse 1996; Gould and Simanis 1997). From studies in animal cells, it is unclear whether cleavage furrow positioning, formation, and contraction are separable events or part of one continuous process. However, genetic analysis in S. pombe has allowed the process of cytokinesis to be divided into several distinct steps. Upon entry into mitosis there is a dramatic rearrangement of the cytoskeleton. The cytoplasmic microtubule arrays depolymerize and reorganize into a mitotic spindle. During this time, Mid1p, Pom1p, and Plo1p function to determine the position at which the medial ring will form (Chang et al. 1996; Sohrmann et al. 1996; Bähler and Pringle 1998; Bähler et al. 1998a), and then the medial ring assembles in the middle of the cell. Many genes have been identified that are required for medial ring formation, most encoding structural components of the actin cytoskeleton such as cdc8 and myo2 (tropomyosin and myosin, respectively) (Balasubramanian et al. 1992; Kitayama et al. 1997; for a summary of medial ring components, see Gould and Simanis 1997). Mutants in these genes cannot assemble medial rings, but do form irregular deposits of septum material. As mitosis progresses, the spindle elongates, carrying one set of chromosomes to each end of the cell, and additional actin structures called patches redistribute from the growing ends of the cell to the medial region adjacent to the medial ring, where they function in deposition of cell wall components (McCollum et al. 1996). At the end of anaphase, the spindle disassembles, and cytoplasmic microtubules begin to regrow from the spindle pole bodies (SPBs)1 at each pole and from the cytoplasmic microtubule organizing centers (MTOCs) at the cell middle (Hagan 1998). In addition, it has been reported that at this time, a microtubule ring forms in the cell middle (Pichova et al. 1995). Also at this time, the medial ring begins to constrict and septal material is deposited behind the constricting ring. Once the septum has formed, the primary septum is degraded, bringing about separation of the daughter cells. Medial ring constriction and septation require the function of at least seven genes, termed the septation initiation genes (sid genes), which include cdc7, cdc11, cdc14 (Nurse et al. 1976; Fankhauser et al. 1995), spg1 (Schmidt et al. 1997), sid1, sid2, and sid4 (Balasubramanian et al. 1998). At the restrictive temperature, these mutants assemble medial rings and redistribute actin patches to the medial region, but then fail to constrict the ring or deposit any septal material (Fankhauser et al. 1995; Balasubramanian et al. 1998). Growth and nuclear division cycles continue in these mutants and the cells eventually lyse after becoming long and multinucleate.
The sequence identities of the sid gene products as well as genetic interactions between them have led to the hypothesis that these genes function in a novel signaling cascade that regulates medial ring constriction and septation (Balasubramanian et al. 1998). The cdc7, sid2, and sid1 genes encode protein kinases (Fankhauser and Simanis 1994; Balasubramanian et al. 1998; McCollum, D., unpublished observations). The spg1 gene encodes a small GTPase in the ras superfamily (Schmidt et al. 1997). The Spg1p GTPase localizes to the SPBs throughout the cell cycle. In interphase cells, Spg1p is in the GDP-bound form, but upon entry into mitosis it converts to the GTP-bound form. Spg1p is then present at both SPBs in the GTP-bound form until anaphase B, when it converts to the guanosine diphosphate (GDP)-bound form at one of the two SPBs. Cdc7p only binds to the GTP-bound form of Spg1p and it only localizes to the SPB(s) when Spg1p is in its active (GTP-bound) form (Sohrmann et al. 1998). Although it has been shown that the Byr4p and Cdc16p proteins function together as the GTPase-activating protein (GAP) for Spg1p (Furge et al. 1998), the guanine nucleotide exchange factor (GEF) has not yet been identified. Overexpression of spg1 will induce septation to occur at any point in the cell cycle, and this phenotype requires the function of Cdc7p, Cdc14p, Cdc15p, and Sid4p (Schmidt et al. 1997; Balasubramanian et al. 1998). These data indicate that Cdc7p and Spg1p may function together in a signaling cascade that initiates at the SPB. It is less clear how the signal to initiate ring constriction is transmitted from the SPB to the medial ring.
The focus of this study is the role of the Sid2p kinase in initiating medial ring constriction and septation. Sequence comparisons suggest that Sid2p may be a homologue of the budding yeast gene Dbf2p (Balasubramanian et al. 1998). Here, we show that Sid2p localizes to the SPBs throughout the cell cycle, and transiently to the cell division site during medial ring constriction and septation. Like Dbf2p (Toyn and Johnston 1994), Sid2p kinase activity peaks at the end of anaphase at the time of septation. Combined cytological and biochemical studies indicate that Sid2p localization and kinase activity depend on the other septation initiation genes, cdc7, cdc11, cdc14, sid1, spg1, and sid4. Thus, it appears that Sid2p kinase may function at a late step of a novel signaling cascade by carrying the signal to initiate division from the spindle poles to the medial ring.
Materials and Methods
Yeast Methods and Strains
The S. pombe strains used in this study are listed in Table . All strains are isogenic to 972 (Leupold 1970). Fission yeast media, growth conditions, and manipulations were carried out as described previously (Moreno et al. 1991). Except where noted, cells were grown in YE medium. Unless otherwise indicated, all experiments involving temperature-sensitive strains were done at a permissive temperature of 25°C and a restrictive temperature of 36°C. Standard genetic and recombinant DNA methods (Sambrook et al. 1989; Moreno et al. 1991) were used except where noted. S. pombe transformations were carried out using either a lithium acetate method (Keeney and Boeke 1994) or electroporation (Prentice 1992). DNA was prepared from bacteria and isolated from agarose gels using Qiagen kits, and from yeast cells as described by Hoffman and Winston 1987. DNA sequencing was performed at the University of Massachusetts Medical School's Nucleic Acid Facility. Oligonucleotide primers were obtained from Integrated DNA Technologies or from Operon Inc.
Table 1.
Schizosaccharomyces pombe Strains Used
| Strain | Genotype | Source |
|---|---|---|
| YDM2 | cdc15-140 ura4-D18 h + | P. Nurse Lab |
| YDM11 | nda3-KM311 leu1-32 h + | M. Yanagida Lab |
| YDM34 | cdc3-124 ura4-D18 h − | P. Nurse Lab |
| YDM38 | sid3-106 ura4-D18 leu1-32 h − | Our stock |
| YDM71 | sid2-250 ura4-D18 leu1-32 h − | Our stock |
| YDM75 | sid1-239 ura4-D18 leu1-32 h + | Our stock |
| YDM105 | leu1-32 ura4-D18 ade6-M210 h − | K. Gould Lab |
| YDM108 | leu1-32 ura4-D18 ade6-216 h + | K. Gould Lab |
| YDM115 | sid4-A1 ura4-D18 leu1-32 h + | Our stock |
| YDM152 | cdc25-22 ura4-D18 h + | P. Nurse Lab |
| YDM272 | cdc14-118 ura4-D18 ade6-M210 h − | P. Nurse Lab |
| YDM273 | cdc7-24 ura4-D18 leu1-32 ade6-M210 h − | Our stock |
| YDM274 | cdc11-123 ura4-D18 h + | P. Nurse Lab |
| YDM415 | sid2-GFP::ura4+ ura4-D18 leu1-32 ade6-M210 h − | This study |
| YDM416 | sid2-GFP::ura4+ ura4-D18 leu1-32 ade6-210 h + | This study |
| YDM419 | sid2-GPF::ura4+ sid4-A1 ura4-D18 leu1-32 h − | This study |
| YDM420 | sid2-GPF::ura4+ sid1-239 ura4-D18 leu1-32 h + | This study |
| YDM421 | sid2-GFP::ura4+ cdc11-123 ura4-D18 h − | This study |
| YDM422 | sid2-GFP::ura4+ cdc7-24 ura4-D18 leu1-32 h − | This study |
| YDM423 | sid2-GFP::ura4+ cdc14-118 ura4-D18 h + | This study |
| YDM425 | sid2-GFP::ura4+ cdc15-140 ura4-D18 h− | This study |
| YDM426 | sid2-GFP::ura4+ nda3-km311 ura4-D18 leu1-32 h− | This study |
| YDM427 | sid2-GFP::ura4+ cdc3-124 ura4-D18 h+ | This study |
| YDM431 | sid2-GFP::ura4+ sid3-106 ura4-D18 leu1-32 h− | This study |
| YDM434 | sid2-GFP::ura4+ sid1-125 ura4-D18 leu1-32 h− | This study |
| YDM435 | sid1-125 ura4-D18 leu1-32 ade6-210 h + | Our stock |
| YDM453 | mad2::ura4+ nda3-km311 ura4-D18 leu1-32 ade6-216 h + | S. Sazer |
| YDM468 | sid2+/sid2::Kan ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-210/ade6-216 h +/h− | This study |
| YDM470 | sid2::Kan ura4-D18 leu1-32 h− plasmid pDM264 | This study |
| YDM471 | cdc7-HA::ura4+ ura4-D18 h− | V. Simanis Lab |
| YDM497 | sid2-13Myc::Kan ura4-D18 leu1-32 h− | This study |
| YDM500 | sid2-13Myc::Kan cdc25-22 ura4-D18 h + | This study |
| YDM508 | sid2-13Myc::Kan cdc7-24 ura4-D18 leu1-32 h + | This study |
| YDM509 | sid2-13Myc::Kan cdc14-118 ura4-D18 h− | This study |
| YDM510 | sid2-13Myc::Kan sid1-125 ura4-D18 leu1-32 h− | This study |
| YDM511 | sid2-13Myc::Kan cdc11-123 ura4-D18 h + | This study |
| YDM512 | sid2-13Myc::Kan sid1-239 ura4-D18 leu1-32 h− | This study |
| YDM513 | sid2-13Myc::Kan sid4-A1 ura4-D18 leu1-32 h− | This study |
| YDM514 | sid2-13Myc::Kan ura4-D18 leu1-32 h + | This study |
| YDM515 | sid2-13Myc::Kan sid3-106 ura4-D18 leu1-32 h + | This study |
| YDM533 | sid2-250 cdc7-HA::ura4+ ura4-D18 leu1-32 h + | This study |
| YDM541 | mad2::ura4+ nda3-km311 sid2-GFP ura4-D18 leu1-32 h + | This study |
| YDM551 | nmt1-spg1::leu leu1-32 ura4-D18 h + | K. Gould Lab |
| YDM554 | nmt1-spg1::leu sid2-250 leu1-32 ura4-D18 h + | This study |
| YDM591 | spg1-B8 h− | V. Simanis Lab |
| YDM614 | sid2-GFP::ura4+ spg1-B8 ura4-D18 h + | This study |
| YDM668 | sid2-13Myc::Kan cdc 15-150 ura4-D18 h + | This study |
Construction of sid2 Deletion and Epitope-tagged Strains
A sid2-null mutant strain was constructed by direct chromosomal integration into a diploid strain of a fragment generated by PCR using plasmid pFA6a-kanMX6 as template, as described (Bähler et al. 1998b). The two primers had 75-bp tails corresponding to the regions immediately 5′ to the sid2 start codon and immediately 3′ of the sid2 stop codon. The PCR fragment was gel purified and transformed into the diploid strain (YDM105 crossed with YDM108) using the lithium acetate method. Transformants were selected on YE G418 plates. PCR analysis of DNA prepared from individual transformants identified a strain bearing the sid2 deletion (YDM468). The sid2 deletion strain was sporulated and tetrads were dissected, which resulted in only two viable G418-sensitive progeny per tetrad. The sid2 deletion strain was transformed with a sid2–green fluorescent protein (GFP) construct (described below) by electroporation. A haploid-null strain (YDM470) bearing an episomal copy of sid2-GFP was obtained by plating a spore preparation from YDM468 expressing sid2-GFP, and selecting for G418-resistant and Ura+ haploid colonies. This strain was grown under conditions that did not select for the plasmid, allowing the plasmid to be lost and the null phenotype examined. Cells that had lost the sid2-GFP plasmid had no GFP signal (see below) and became long and multinucleate (data not shown). Costaining with antibodies against tubulin, as well as Cdc4p and Arp3p, which stain the medial ring and actin patches, respectively, also showed no readily apparent abnormalities in these structures (data not shown). Thus, the sid2-null phenotype was indistinguishable from that of the temperature-sensitive allele of sid2, sid2-250 (Balasubramanian et al. 1998).
A strain expressing a Sid2p-GFP COOH-terminal fusion was prepared by first amplifying the last 800 bp of the sid2 gene by PCR. The 5′ primer contained an Xba1 site followed by an in frame stop codon, and the 3′ primer contained an Nde1 and Kpn1 site. The PCR product was directionally cloned into the integrating vector pJK210 (Keeney and Boeke 1994), using the Xba1 and Kpn1 sites from the primers. The GFP (S65T) DNA fragment was cloned in frame at the 3′ end of the sid2 gene as an NdeI fragment to yield plasmid pDM238. The final construct was linearized in the sid2 coding sequence using EcoR1 and transformed into yeast (YDM105). Note that the integration of this plasmid results in the full-length gene fused to the GFP tag, as well as an untagged COOH-terminal fragment of the sid2 gene which is not expressed due to the in frame stop codon at the 5′ end of this fragment. Integrants were selected on ura(−) plates, and correct integration was confirmed by PCR of integrant genomic DNA. The final strains (YDM415/YDM416) were then crossed to various mutant strains, producing the strains listed in Table (YDM420–YDM442, YDM541, YDM614). Sid2p was tagged at its COOH terminus with 13 tandem copies of the myc epitope (13Myc) by direct chromosomal integration into strain YDM105 of a fragment generated by PCR using plasmid pFA6a-13Myc-kanMX6 as template (Bähler et al. 1998b). The two PCR primers had 20 bp homologous to the DNA template followed by 75-bp tails corresponding to the regions 5′ and 3′ of the sid2 stop codon. The resulting strain (YDM497 or YDM514) was checked for correct integration by PCR. This strain was then crossed with various mutant strains, producing strains listed in Table (YDM500-515, YDM545). All sid2 epitope–tagged strains were indistinguishable from wild-type with respect to growth rates and cellular morphology.
Construction of sid2-containing Plasmids
A number of different plasmids were constructed for expressing sid2 or tagged versions of sid2 in S. pombe cells. Plasmid pDM264 allows for expression of sid2 from its own promoter with a COOH-terminal GFP fusion in the vector pUR19 (Barbet et al. 1992). Plasmid pDM264 was constructed by starting with the sid2 genomic clone, pDM99, in the vector pUR19, and then replacing the COOH-terminal fragment of sid2 between the Nhe1 site in the sid2 coding region, and the Kpn1 site in the 3′ flanking region, with the Nhe1 to Kpn1 fragment of plasmid pDM238 (see above), which contains the COOH terminus of sid2 fused with GFP. Several other plasmid vectors were constructed for expression of sid2 or a kinase-dead version of sid2 (see below), either alone or as NH2-terminal GFP or triple hemagglutinin (HA) epitope fusions from the thiamine-repressible promoter in vector pRep42 (Basi et al. 1993). The sid2 gene or a kinase-dead version, sid2-K238R (see below), were amplified by PCR using oligos that added Ase1 and EcoICR1 sites at the NH2 and COOH termini, respectively. These fragments were then cloned into the Nde1/Sma1 cut vectors pRep42 (Basi et al. 1993), pRep42GFP, and pRep42HA (Craven et al. 1998). Constructs bearing the nmt1 promoter (Maundrell 1990) were regulated by the addition of thiamine to a final concentration of 2 μM.
Site-directed mutagenesis was employed to create a point mutation in the sid2 gene at a site that would alter the proposed ATP-binding domain of the kinase (K238 to R). A sid2 genomic clone in the plasmid pUR19 was mutated by a PCR-based Quikchange mutagenesis system (Stratagene), resulting in the plasmid pDM343. The presence of the K238R mutation was confirmed by DNA sequencing.
Microscopy
Indirect immunofluorescence staining was performed as described previously (Balasubramanian et al. 1997). The tubulin antibody TAT-1 (Woods et al. 1989) was a generous gift from K. Gull (University of Manchester, Manchester, UK). Cdc4p and Arp3 antibodies from our lab stock were used at a 1:100 dilution. HA antibodies (BABCO) were diluted 1:200. Primary antibodies were detected with anti–rabbit or anti–mouse Texas red or FITC-IgG (Molecular Probes). DNA was visualized with DAPI (Sigma Chemical Co.) at 20 μg/ml. Images were captured using a Nikon Eclipse E600 microscope with a cooled CCD camera (Dage 300; MVI) and IPlab Spectrum software (Signal Analytics Corp.).
Immunoelectron microscopy was done as described in Ding et al. 1997 with antibodies against GFP (a generous gift of P. Silver, Harvard Medical School, Boston, MA) or the Myc epitope (a generous gift of H. McDonald, Vanderbilt University, Nashville, TN).
Immunoprecipitations and Western Blot Analysis
Protein lysates were prepared from 1.0–2.5 × 109 cells, which were collected by centrifugation and frozen on dry ice. All subsequent manipulations were carried out at 4°C or on ice. Cells in NP-40 buffer (1% NP-40, 150 mM NaCl, 2 mM EDTA, 6 mM NA2HPO4, 4 mM NaH2PO4, 0.15 μg/ml PMSF, 5 μg/ml each aprotinin, leupeptin, and pepstatin) were lysed by vortexing vigorously with glass beads (Sigma Chemical Co.). Protein lysates were prepared as described in Moreno et al. 1991. Relative protein concentrations were determined using a Coomassie assay (BioRad).
Immunoprecipitations were carried out by adding to the NP-40 cell lysates either 1 μl of anti-myc 9E mouse monoclonal IgG (M. Jacobs, Tufts University, Boston, MA) or 1 μl anti-HA mouse monoclonal IgG (H. McDonald, Vanderbilt University) as appropriate, followed by incubation for 1 h on ice. Immune complexes were purified by adding 25 μl of a 1:1 slurry of protein G–Sepharose beads (Amersham Pharmacia Biotech), incubating for 30 min on a rocker at 4°C, and pelleting bound material by centrifugation in a microfuge for 1 min. Beads were washed three times with 1 ml NP-40 buffer.
For detection of Myc or GFP epitope–tagged Sid2p in total protein lysates, NP-40 cell lysates (described above) were separated by SDS-PAGE (7%), and transferred to Immobilon P nylon (Millipore Corp.) using a semidry blotting apparatus (Owl Scientific). Blots were probed with the anti-myc IgG or anti-GFP IgG (Clontech) at a 1:1,000 dilution, and developed using an alkaline phosphatase chemiluminescent system (BioRad).
In Vitro Kinase Assays
Immune complex bead preparations were washed once in 1 ml of kinase assay buffer (25 mM MOPS, pH 7.2, 60 mM β-glycerol phosphate, 15 mM MgCl2, 15 mM p-nitrophenylphosphate, 1 mM DTT, 0.1 mM sodium vanadate, 1% NP-40, 50 μM ATP (unlabeled), 5 μg/ml each aprotinin, leupeptin, and pepstatin). Washed immunoprecipitates were incubated at 30°C for 30 min in 20 μl kinase assay buffer with 10 μg myelin basic protein (MBP) or histone (Sigma Chemical Co.), and 2 μl of a 1:10 dilution of 10 mCi/ml (3,000 Ci/mmole) [32P]ATP (AA0068; Amersham Pharmacia Biotech). Reactions were stopped with the addition of SDS-PAGE sample buffer and half of each sample was resolved on 15% polyacrylamide gels. Gels were dried and imaged on a PhosphorImager (Molecular Dynamics). The other half of each sample was probed for Sid2p-13Myc or HA-Sid2p by Western blotting, and used to normalize Sid2p kinase activity. For each experiment, a cell lysate from cells that do not contain Sid2p-13Myc or HA-Sid2p was used as a control, and the amount of MBP phosphorylation observed was used to determine the amount of background for each experiment.
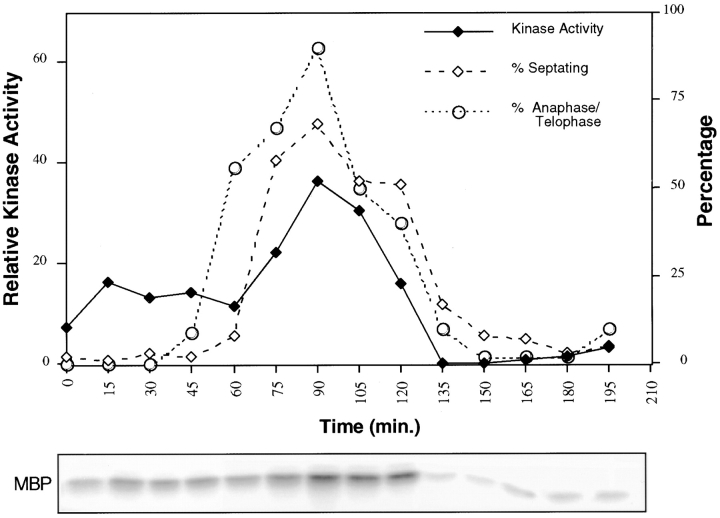
For the cell cycle profile of kinase activity, strain YDM500 (cdc25-22, sid2-13Myc) was grown to log phase at 25°C, shifted to 36°C for 3 h, then shifted back to 25°C. Cells were collected every 15 min for 4–5 h. At each time point, a 50 ml culture of cells was pelleted and frozen in a dry ice/ethanol bath, a sample of the cells was fixed in −20°C methanol and stained with DAPI, and a sample of the cells was examined by phase–contrast microscopy for the appearance of a septum. Once all time points were collected, cell pellets were assayed for in vitro kinase activity as described above.
Results
The Sid2p Kinase Is Localized to the SPBs and the Cell Division Site
To better understand how the Sid2p kinase controls initiation of medial ring constriction and septation, we sought to determine the localization of the protein in S. pombe cells. A strain was constructed that expressed a Sid2p-GFP fusion from the normal chromosomal locus (see Materials and Methods). In interphase cells, a single spot of GFP fluorescence was observed at the periphery of the nucleus, whereas two spots of GFP fluorescence were seen in mitotic cells (Fig. 1, A–C). At the end of anaphase, Sid2p appeared as a band in the medial region as well as two nuclear-associated dots (Fig. 1 A, panels 3 and 4; Fig. 1 B, bottom panel). This band of Sid2p colocalized with the medial ring during initiation of ring constriction (Fig. 1 B, bottom panel). As the ring constricted and the primary septum formed, Sid2p localized along either side of the developing septum (Fig. 1 A, panel 4). Following septum formation, but before cell separation, Sid2p disappeared from the middle of the cell (Fig. 1 A, panel 5). Real-time examination of living cells stained with calcofluor showed that Sid2p localized to the medial region just before the appearance of septal material (Sparks, C., S. Wheatley, and Y.L. Wang, unpublished observations).
Figure 1.
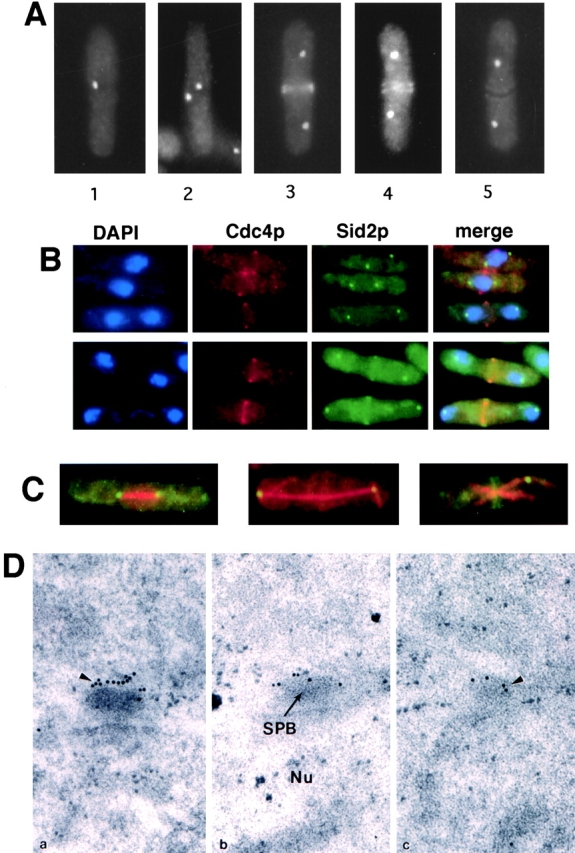
Sid2p-GFP localizes to the SPB and the site of cell division. Sid2p-GFP–expressing cells (YDM415) were fixed and photographed at various stages as described in the text (A), or were fixed and stained for DNA (DAPI), and Cdc4p (B) or tubulin (C). (B) DAPI, Cdc4p, Sid2p-GFP, and merged images are shown. Mitotic cells that had not yet completed anaphase show Cdc4p medial rings but no medial Sid2p-GFP signal (top panel). Postanaphase cells show colocalization between Cdc4p and Sid2p-GFP (bottom panel). (C) Sid2p-GFP (green/yellow) images were merged with the tubulin images (red). Metaphase (left panel), late anaphase (middle panel), or telophase (right panel) cells are shown. (D) Sid2p-GFP localizes to the SPB by immunoelectron microscopy. Cells were grown at 32°C to mid-log phase and prepared for immunoelectron microscopy with antibodies against GFP and with gold-labeled secondary antibodies. Three serially sectioned images are shown. Arrowheads indicate gold particles labeling the SPB, and the position of the nuclear matrix (Nu) and the SPB is indicated.
The nuclear-associated spot-like localization of Sid2p suggested that Sid2p was a component of the SPB. To confirm SPB localization, we fixed and stained the Sid2p-GFP–expressing cells with anti-tubulin antibody. Sid2p was localized at the ends of the mitotic spindle in cells undergoing mitosis, consistent with its localization to the SPB (Fig. 1 C). To unequivocally establish that Sid2p was a component of the SPB, as well as to assess if Sid2p localized to the nuclear or cytoplasmic faces of the SPB, Sid2p localization was examined using strains expressing Sid2p-GFP by immunoelectron microscopy. As shown in Fig. 1 D, the SPB appears as an electron-dense disk-shaped structure at the edge of the nucleus by electron microscopy. Using GFP-specific antibodies, we found that Sid2p localized to the outer cytoplasmic face of the SPB (Fig. 1 D). Serial sections through a representative SPB body are shown. The distribution pattern of Sid2p at the SPB was the same in both interphase and mitotic cells (data not shown). Essentially identical results were obtained following electron microscopy when Sid2p-13Myc cells were stained with Myc-specific mAbs (data not shown). Thus, Sid2p is a bona fide component of the SPB and is associated with the cytoplasmic face of the SPB. Sid2p was not detected at the division site in septating cells using either Myc- or GFP-specific antibodies. This may be due to inaccessibility of Sid2p to antibodies when it is at the division site, since we were also unable to observe Sid2p staining at the division site by conventional immunofluorescence using either Myc- or GFP-specific antibodies (data not shown).
Sid2p Depends on the Medial Ring and Microtubules to Localize to the Cell Division Site
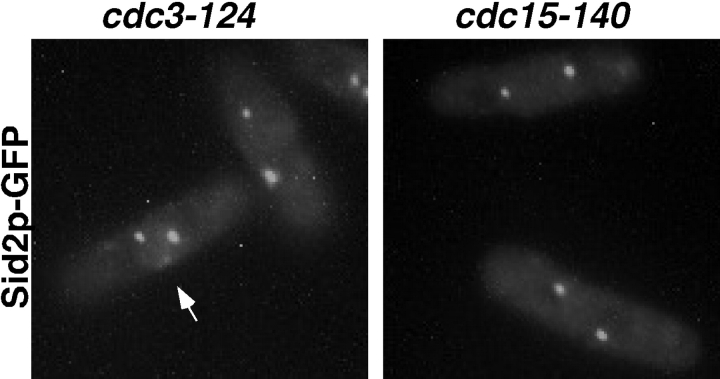
To determine if localization of Sid2p to the middle of the cell required the medial ring, we examined its localization in two different types of medial ring mutants. In cdc3 mutants, medial rings are not formed (Balasubramanian et al. 1994) and septal material is deposited in irregular patches on the cell cortex. We found that in cdc3 mutants, Sid2p never formed the medial band, but faint patches of Sid2-GFP were occasionally observed at the cell cortex (Fig. 2, arrow). Another type of medial ring mutant examined was a cdc15 mutant. In these cells, an actin-containing medial ring will form in the middle of the cell (Balasubramanian et al. 1998) but these rings lack the Cdc15p component (Fankhauser et al. 1995). Interestingly, we could not detect any Sid2p at the medial cortex in cdc15-140 mutants (Fig. 2, right panel). In both cases >200 cells were examined. These observations indicate that Sid2p requires the actin ring for its localization to the medial cortex, and that there may be an interaction between Cdc15p and Sid2p at the medial ring.
Figure 2.
Localization of Sid2p-GFP to the cell division site depends on the presence of an actin ring and Cdc15p. Both cdc3-124 (YDM427) and cdc15-140 (YDM425) cells expressing Sid2p-GFP were grown to mid-log phase at 25°C, shifted to 36°C for 2.5 h, then fixed and photographed. Arrow indicates structure described in the text.
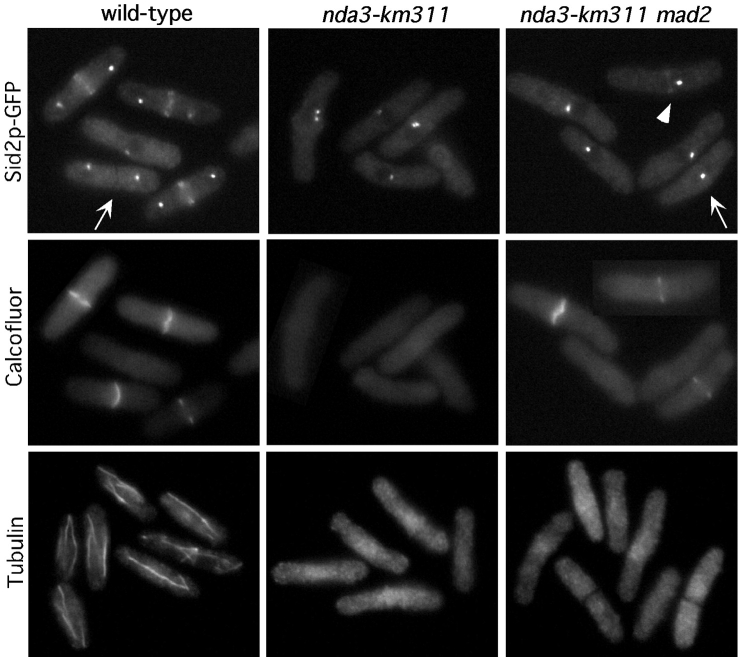
Next, we asked whether microtubules were required for the redistribution of the Sid2p kinase from the SPB to the division site. We began by examining the distribution of Sid2p-GFP in the cold-sensitive tubulin mutant strain nda3-KM311. After 4 h at the restrictive temperature of 19°C, 70% of cells had arrested in mitosis with condensed chromosomes, and no cells had septa (Fig. 3, and data not shown). Also, no intact microtubules were detected in these cells by indirect immunofluorescence microscopy (Fig. 3). We found that Sid2p remained at the SPB in these cells (Fig. 3). Thus, the microtubule cytoskeleton is not required to maintain the localization of Sid2p to the SPB. At the block-point, nda3-KM311 mutants undergo mitotic arrest with fully formed medial rings (Chang et al. 1996). Sid2p failed to accumulate at the medial rings in cold-arrested nda3-KM311 cells, suggesting that an intact microtubule cytoskeleton, perhaps in the form of the postanaphase array, was required for accumulation of Sid2p at the division site.
Figure 3.
Sid2p-GFP requires microtubules to efficiently localize to the division site. Sid2p-GFP–expressing cells, or Sid2p-GFP cells carrying either the nda3-km311 mutation (YDM426), or the nda3-km311 and mad2Δ (YDM541) mutations were grown to mid-log phase at 30°C and then shifted to 19°C for 4 h and fixed in the cold, and either stained with calcofluor, or stained for tubulin. Images were captured for each strain showing the GFP signal (Sid2p-GFP) and calcofluor staining (Calcofluor), or the tubulin staining. A representative montage of cells is shown. Arrows and arrowheads indicate structures described in the text.
There are several possible explanations for why Sid2p failed to accumulate at the medial rings formed in the tubulin mutant cells. Sid2p could be transported from the SPB to the cleavage site down cytoplasmic microtubules, or alternatively, the medial ring of microtubules (Pichova et al. 1995) may be required to maintain Sid2p at the division site. Another distinct possibility is that these cells have arrested in metaphase, before the end of anaphase when activation of the pathway to signal the initiation of cytokinesis is presumably turned on. Of course, it is also possible that Sid2p localization to the cell middle requires both activation of the signaling pathway and the presence of intact microtubules. To test between these possibilities, we examined the Sid2p distribution in a strain carrying a deletion of the spindle checkpoint gene, mad2, in addition to the nda3 tubulin mutation. These cells do not arrest in metaphase, but instead continue to cycle without any microtubules present, eventually undergoing cytokinesis and cleaving through unsegregated chromosomes (He et al. 1997). As expected, we found that unlike the nda3 mutant alone, these cells did not remain blocked in mitosis. Out of >200 cells counted, only 7% had condensed chromosomes, 30% of the population had septum staining, and like the nda3 mutant cells, these cells had no intact microtubules by immunofluorescence after 4 h at 19°C (Fig. 3). Upon initial examination of these cells, it appeared that although 30% of the cells displayed septum staining, Sid2p was not localized to the medial region of these cells. However, careful scrutiny of these cells showed that a very faint Sid2p signal could be observed in the medial region of 5% of the septated cells. The faint Sid2p medial signal was always observed in cells that had not completed septum formation (Fig. 3, arrowhead), although not all septating cells had an obvious Sid2p signal in the medial region (Fig. 3, arrow). It is not surprising that Sid2p was not observed in the medial region in the majority of the cells that displayed septum staining, since most of these cells had completed septum formation, and in wild-type cells, Sid2p normally disappears from the medial region at that time (Fig. 3, arrow). Quantitation of the fluorescence intensity of the medial staining in 20 wild-type and 20 of the nda3 mad2 cells in which a medial Sid2p-GFP signal could be visualized showed that the Sid2p-GFP signal was ∼10-fold less intense in the nda3 mad2 cells than that observed in wild-type cells. The simplest explanation for these results is that normal Sid2p localization to the medial ring requires both anaphase-induced activation of the signaling pathway and the presence of intact microtubules. In the nda3 mad2 mutant cells, enough activated Sid2p may be able to diffuse to the medial ring, causing septation to initiate, but Sid2p is unable to accumulate to the levels seen in wild-type cells in the absence of microtubules.
Sid2p Depends on the Septation Initiation Gene Products to Distribute Properly
It has been proposed that the Sid groups of proteins (Cdc7p, Cdc11p, Cdc14p, Sid1p, Sid2p, Spg1p, and Sid4p) function as a part of a novel signal transduction cascade (Balasubramanian et al. 1998). Thus, in the following sections we describe several lines of experimentation designed to test if Sid2p functions in a signaling cascade, and if so, where it functions with respect to the other Sid proteins. As one way to assess the order of function for these genes, we asked if any of the other sid genes were required for the distinctive distribution pattern of Sid2p. We found that Sid2p-GFP could not localize to both the SPB and the medial region in sid4, cdc7, and cdc11 mutants (Fig. 4 A, and data not shown). In sid1, spg1, and cdc14 mutants, Sid2p-GFP localized to the SPB but not to the medial region (Fig. 4 A, and data not shown). In all cases, the Sid2p localization pattern was examined in at least 200 cells. These results are consistent with the possibility that sid2 functions downstream of all of the other known genes in this signaling cascade. We also checked Sid2p-GFP protein levels in the mutant strains in which the Sid2p-GFP signal was lost from the SPB at the restrictive temperature, to test whether Sid2p was being degraded or simply displaced in these mutant cells. We found that the Sid2p protein levels were similar to wild-type cell levels (Fig. 4 B) and, therefore, the Sid2p appears to be blocked from localizing properly in the mutant backgrounds. Thus, the localization of Sid2p to the division site requires the function of all of the Sid proteins. In addition, Cdc7p, Cdc11p, and Sid4p are also required for its localization to the SPB.
Figure 4.
Sid2p-GFP localization depends on the other Sid proteins. (A) sid1-239 (YDM420) and cdc11-123 (YDM421) cells expressing Sidp2-GFP were grown to mid-log phase at 25°C and then shifted to 36°C for 2.5 h, and fixed and stained for DNA. (B) Sid2p-GFP protein is stable in the sid mutants. Cells expressing no Sid2p-GFP (Wild-type; YDM105), or expressing Sid2p-GFP in a wild-type background (sid2-GFP; YDM415) or in sid4-A1 (sid2-GFP, sid4-A1; YDM419), sid1-239 (sid2-GFP, sid1-239; YDM420), cdc11-123 (sid2-GFP, cdc11-123; YDM421), or cdc7-24 (sid2-GFP, cdc11-123; YDM422) mutant background were grown as described in A, and then Sid2p-GFP levels were analyzed by Western blotting with antibodies against GFP.
Spg1p and Cdc7p Are Upstream of Sid2p in the Signaling Pathway, Initiating Actin Ring Constriction and Septation
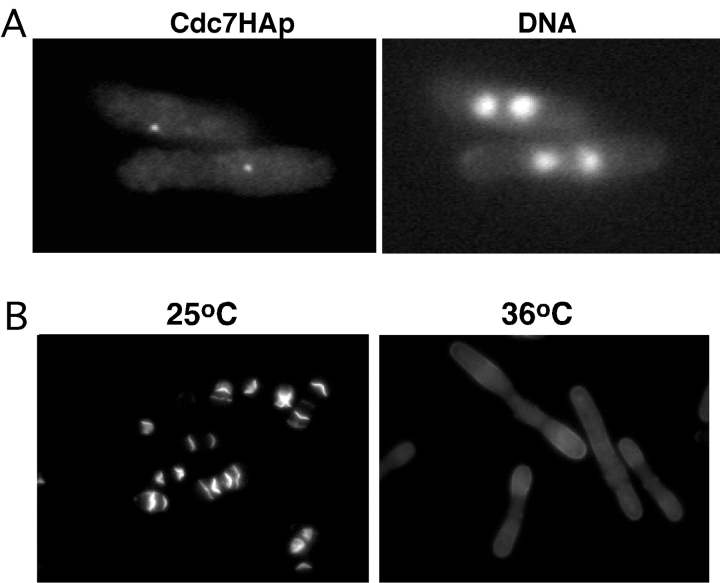
A previous study has shown that Cdc7p functions downstream of Spg1p (Sohrmann et al. 1998). The Sid2p localization experiments described above suggest that Sid2p may function downstream of both Spg1p and Cdc7p. To test this hypothesis in another way, we asked whether Cdc7p depends on Sid2p for its proper distribution pattern. We examined the localization of an HA epitope–tagged Cdc7p (a gift from V. Simanis, ISREL, Epalinges, Switzerland) in a sid2-250 mutant. Cells grown at the permissive temperature, or incubated at the restrictive temperature for 3 h, were fixed and stained with HA antibodies. At permissive temperature, 28% (61/219) of cells examined displayed Cdc7p staining at the SPB. At the restrictive temperature, 53% (156/294) of cells showed localization of Cdc7p to one or more SPBs (Fig. 5 A). The increased percentage of cells with Cdc7p SPB staining at the restrictive temperature reflects the fact that these cells had become multinucleate. Therefore, Cdc7p does not require the function of Sid2p to localize properly, suggesting that Cdc7p is upstream of Sid2p in this cascade.
Figure 5.
Sid2p functions downstream from Cdc7p and Spg1p. (A) sid2-250 cdc7-HA cells (YDM533) were grown to mid-log phase at 25°C and then shifted to 36°C for 2.5 h, and fixed and double stained for Cdc7p using antibodies against the HA epitope or for DNA. (B) sid2-250 nmt1-spg1 cells (YDM554) were grown to mid-log phase at 25°C in medium containing thiamine, and then the culture was shifted into medium lacking thiamine and grown at 25°C for an additional 23 h. The culture was then split two ways into medium lacking thiamine, with one culture placed at 25°C and the other at 36°C for 5 h, and then cells were fixed and stained with calcofluor.
Cells that overexpress spg1 undergo multiple rounds of septation but never complete cell separation (Schmidt et al. 1997). If Sid2p functions downstream of Cdc7p, then it should be downstream of Spg1p as well. To test this, we asked if the sid2-250 phenotype was epistatic to the Spg1p overproduction phenotype. We examined the consequences of overexpressing the spg1 gene using the thiamine-repressible nmt1 promoter in the sid2-250 mutant. Overexpression of spg1 at the permissive temperature caused cells to form multiple septa (Fig. 5 B) as reported previously in wild-type cells (Schmidt et al. 1997). Of 100 cells counted, 98 had 2 or more septa. However, at restrictive temperature, out of 100 cells counted, none of the cells formed septa, and these cells instead became long and multinucleate like sid2-250 mutants (Fig. 5 B), demonstrating that the sid2-250 phenotype is epistatic to the spg1 overexpression phenotype. Together, these experiments are consistent with Sid2p functioning downstream of Cdc7p and Spg1p in the pathway to initiate actin ring constriction and septation.
Sid2p Acts as a Kinase In Vitro
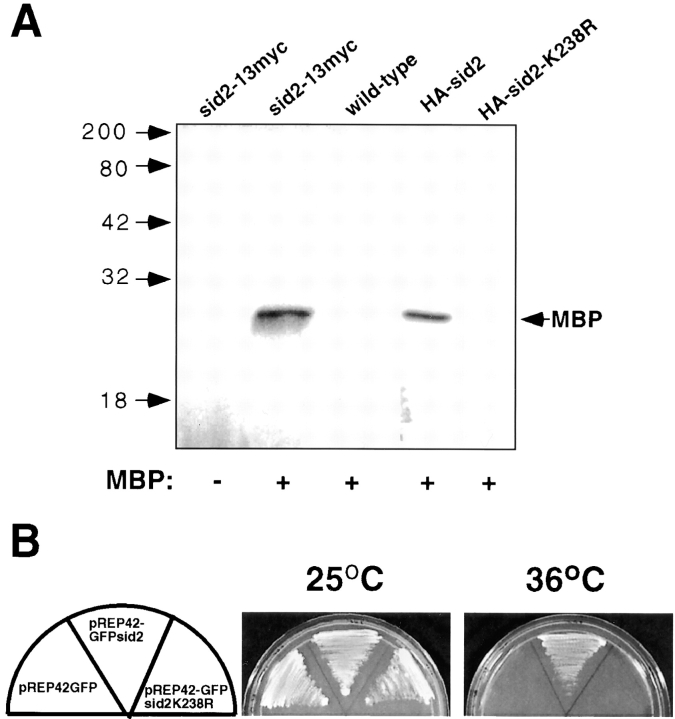
Sequence analysis of the sid2 gene predicts that the protein functions as a protein kinase (Balasubramanian et al. 1998). To test if Sid2p had kinase activity in vitro, we used a strain which expressed a 13Myc epitope tag inserted in frame at the 3′ end of the chromosomal sid2 gene (see Materials and Methods). Sid2p immune complexes were prepared from these cells using anti-myc antibodies and assayed for kinase activity using MBP as an artificial substrate. Sid2p was found to efficiently phosphorylate MBP (Fig. 6 A). Appreciable nonspecific kinase activity in cells not expressing Sid2p-13Myc was not detected (Fig. 6 A, wild-type). We tested the ability of the Sid2p to autophosphorylate by eliminating the substrate in the assay, however no autophosphorylation was detected. To ensure that the kinase activity that we were measuring was due to Sid2p and not an associated kinase, we created an episomally expressed HA-tagged kinase-dead version of the protein by introducing a point mutation in the proposed ATP binding site of the kinase that changed lysine 238 to arginine (K238R) (see below). Both HA-Sid2p and HA-Sid2-K238R were expressed in wild-type cells and their kinase activity was measured. This showed that although HA-Sid2p did display kinase activity, the kinase-dead version HA-Sid2-K238R did not (Fig. 6 A).
Figure 6.
Sid2p has protein kinase activity which is essential for its function. (A) Immune complex kinase assays were performed using myc antibodies (see Materials and Methods) on lysates from wild-type cells, or cells expressing Sid2p-13Myc, HA-Sid2, or HA-Sid2-K238R. Assays were carried out with (+) or without (−) addition of MBP as an artificial substrate. (B) sid2-250 cells (YDM71) transformed with plasmids pRep42, pRep42-GFPsid2, or pRep42-GFPsid2-K238 were plated at 25°C or 36°C on plates containing thiamine.
We also wanted to test the importance of Sid2p kinase activity for the function of the protein in vivo. The K238R mutation was introduced into a plasmid carrying a sid2 genomic clone, or because we suspected that overexpression of the sid2-K238R allele might be toxic, we expressed sid2-K238R in the vector pREP42 under the control of an attenuated version of the thiamine-repressible nmt1 promoter either with or without a GFP or HA tag (see Materials and Methods). The GFP tag allowed us to determine if Sid2p kinase activity was required for proper localization. The sid2-K238R gene expressed off its own promoter was presumably lethal to cells, since no colonies formed after transformation of this plasmid into wild-type cells. The pRep42 sid2-K238R constructs were not lethal when weakly expressed under thiamine repression in wild-type yeast cells. However, when induced, the cells grew very slowly and appeared to be quite sick (data not shown). We then transformed each of these plasmids into the sid2-250 strain to ask if the kinase-dead version of sid2 could rescue the sid2 mutant phenotype at 36°C. Each strain was grown on selectable plates at 25°C, then shifted to 36°C in the presence of thiamine (Fig. 6 B). Only the wild-type version of the sid2 gene could rescue the sid2-250 strain at 36°C. This showed that under these conditions, sufficient GFP-Sid2p (or Sid2p, not shown) was expressed to rescue the sid2-250 mutant. However, the Sid2p-K238R protein was not capable of rescue under the same circumstances. Removing thiamine and thus allowing for the overexpression of the kinase-dead Sid2p results in a lethal phenotype at the permissive temperature (data not shown). Overexpression of wild-type Sid2p had no effect on cell growth or morphology in both wild-type and sid2-250 mutant cells. Both the wild-type and kinase-dead GFP-Sid2p were expressed in wild-type cells and localized properly to the SPBs and the cleavage furrow (data not shown), indicating that Sid2p kinase activity is not required for proper localization of the protein.
Sid2p Kinase Activity Peaks during Actin Ring Constriction and Septation
Next, we were interested in determining if Sid2p kinase activity was cell cycle regulated. To test this, we prepared a strain that expressed Sid2p-13Myc in a cdc25-22 background. When shifted to 36°C, cdc25-22 mutant cells arrest at the G2/M transition. Upon release to the permissive temperature, these cells proceed synchronously through the cell cycle. Thus, the cdc25-22 mutation was used to generate populations of synchronous cells for analysis of Sid2p kinase activity throughout the cell cycle. Cells were collected every 15 min after release from a 36°C block, and Sid2p kinase activity was measured at each time point (Fig. 7). The percentages of cells undergoing anaphase and septation were also determined at each time point (Fig. 7). Interestingly, the Sid2p kinase activity peaked during actin ring constriction and septation at the end of anaphase (Fig. 7). Low levels of kinase activity were also observed earlier in mitosis. The significance of this earlier activity is not clear. However, the majority of the Sid2p kinase activity peaks when Sid2p is accumulating at the medial ring during ring constriction and septation.
Figure 7.
Sid2p kinase activity is cell cycle regulated. cdc25-22 sid2-13Myc cells (YDM500) were blocked at 36°C for 3 h then returned to 25°C, allowing them to synchronously enter the cell cycle. Every 15 min, samples were collected for measurement of kinase activity as well as the percentage of cells that were in anaphase/telophase (circles) and the percentage undergoing septation (open diamonds). Sid2p kinase activity was plotted (filled diamonds) by first normalizing the amount of MBP phosphorylation (bottom panel) to the amount of Sid2-13Myc protein present as determined by Western blotting.
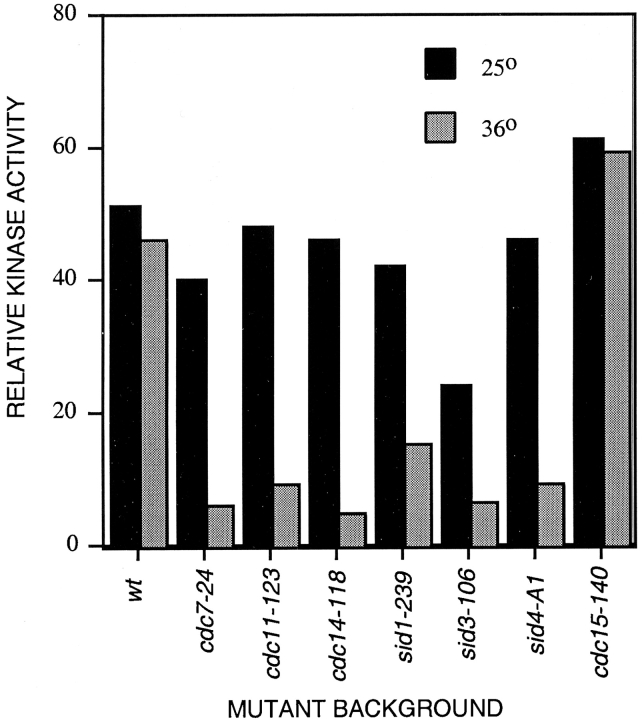
Sid2p Kinase Activity Depends on the Septation Initiation Gene Products but Not Cdc15p
Having demonstrated that the Sid2p localization was dependent on the activity of the other gene products that function in septation and medial ring constriction (Fig. 4), we now wanted to see if these same gene products were also required for Sid2p kinase activity. Sid2p kinase activity was measured using strains expressing Sid2p-13Myc in each of the septation initiation mutant backgrounds, which had been grown at permissive or restrictive temperatures. In all of the mutant backgrounds, Sid2p kinase activity was greatly diminished at restrictive temperature (Fig. 8). The reduced kinase activity of Sid2p in these mutant strains does not appear to be due to a nonspecific cell cycle block, since nuclear division cycles continue in these mutants and they display a mitotic index similar to that of wild-type cells (Nurse et al. 1976; Marks et al. 1992; data not shown). Thus, the Sid2p kinase appears to require the function of the other septation initiation gene products to be active. This would support our other results which place Sid2p at the end of the signaling cascade that regulates medial ring constriction and septation. It could also be proposed that the sid group of genes is only required for Sid2p to localize to the medial ring, and then the protein becomes activated at the ring. To test this, we also examined Sid2p kinase activity in the medial ring mutant cdc15, which is also required for medial Sid2p localization. Sid2p activity was not lost at the restrictive temperature in the cdc15 mutant, suggesting that Sid2p does not have to localize to the medial ring to become activated.
Figure 8.
Sid2p kinase activity depends on the other Sid proteins. Cells expressing Sid2p-13Myc in an otherwise wild-type background (YDM497; wt), or in cdc7-24 (YDM508), cdc11-123 (YDM511), cdc14-118 (YDM509), sid1-239 (YDM512), sid3-106 (YDM515), sid4-A1 (YDM513), or cdc15-140 (YDM668) mutant backgrounds were grown to mid-log phase at 25°C then split into two cultures, one at 25°C and one at 36°C. Each culture was then incubated for 3 h at its respective temperature and assayed for kinase activity. Relative kinase activities are depicted graphically.
Discussion
To maintain genomic stability and proper ploidy, it is crucial that cell division occurs at the end of anaphase after chromosome segregation. Genetic analysis in S. pombe has identified a group of interacting genes, which includes cdc7, cdc11, cdc14, sid1, sid2, sid4, and spg1 (Nurse et al. 1976; Schmidt et al. 1997; Balasubramanian et al. 1998), required to initiate constriction of the medial ring and septation at the end of anaphase. It was shown that the Spg1p GTPase and the Cdc7p kinase both reside at the SPBs (Sohrmann et al. 1998), perhaps to be able to sense that anaphase was complete and then to pass on a signal to initiate cell division. Here we show that Sid2p may function downstream of Spg1p and Cdc7p to pass the signal on to the medial cortex that it is time to divide.
Sid2p Localization
It had been shown previously that Spg1p and Cdc7p localize to the SPBs but not the cell division site (Sohrmann et al. 1998), raising the question of how the signal to divide becomes transmitted from the poles to the division site. Here we show that Sid2p localizes to both the SPBs and to the cell division site at the time of cell division, suggesting that Sid2p may transmit the signal to divide from the poles to the division site. We also identified a number of proteins required for proper localization of Sid2p to both the SPB and the division site. It appears that Sid2p is an actual component of the SPB since it does not require microtubules for localization to the SPB. However, localization of Sid2p to the SPB does require Cdc7p, Cdc11p, and Sid4p. One explanation of these results could be that there is a protein complex between these proteins at the SPB, and if it is disrupted, other components do not localize properly. However, initial coimmunoprecipitation experiments, using mild conditions (1% NP-40 lysis buffer), looking at endogenous protein–protein interactions failed to detect any complex formation between Sid2p and either Cdc7p or Sid4p (Sparks, C., and D. McCollum, unpublished observations), although it is possible that the methods used to prepare the cell lysates did not solubilize or preserve protein complexes at the SPB. A complex between Sid2p and Cdc7p also seems unlikely, since Cdc7p only resides at the SPB during mitosis, whereas Sid2p is always at the SPB. Furthermore, in an spg1-B8 mutant, Sid2p does localize to the SPBs even though Cdc7p does not (Sohrmann et al. 1998), suggesting that Cdc7p may function in the cytoplasm to promote Sid2p localization to the SPB. These results are somewhat surprising and we do not have a simple explanation for them at this time.
Further experiments showed that Sid2p requires microtubules for it to efficiently accumulate at the division site. The fact that the nda3 mad2 cells are able to septate despite poorly localizing Sid2p to the division site poses somewhat of a paradox. Although Sid2p accumulates inefficiently at the division site in the absence of microtubules, the fact that some Sid2p can still localize to the division site suggests that binding interactions between freely diffusing Sid2p and medial ring components are sufficient to localize enough Sid2p to the medial ring to trigger septation. Since Sid2p presumably functions in a catalytic manner, it may not be necessary have large amounts of Sid2p at the division site to initiate septation. Similarly, it has been observed that in budding yeast, myosin rings can form in the absence of actin; however, ring formation is much less efficient, suggesting that transport of myosin, facilitated by F-actin, is important for accumulation of myosin at the medial ring, but in the absence of F-actin, myosin can accumulate by diffusion mechanisms (Bi et al. 1998).
Although cells can clearly septate without the efficient microtubule-mediated accumulation of Sid2p at the division site, this mechanism may be important in normal cells to ensure that cytokinesis initiates precisely at the end of anaphase. It is worth noting that precisely at the time when Sid2p begins to appear at the division site, the spindle is breaking down, the postanaphase array of microtubules is forming, and a medial band of microtubules is present (Pichova et al. 1995). At this time, microtubules can be observed running from the cell division site to the SPBs. It has been proposed that these microtubules function to position the nucleus in the middle of the new cell away from the division site to ensure that the nucleus does not get cut by the septum (Hagan and Yanagida 1997). Given the timing of the appearance of these microtubules, it is tempting to speculate that Sid2p moves along these microtubules from the SPB to the division site. This would serve as an efficient mechanism to couple proper positioning of the nucleus with initiation of cell division. The localization of Sid2p to the outer cytoplasmic face of the SPB places it in a position from which it could move down astral microtubules emanating from the pole. However, it is also possible that the medial band of microtubules is important for Sid2p localization, and at this point we cannot say how microtubules function to help localize Sid2p to the division site. The fact that one of the putative upstream activators of Sid2p, Cdc7p, only localizes to one of the SPBs instead of two, like Sid2p, suggests that if Sid2p moves along microtubules from the SPB to the division site, it may only come from one of the two poles. However, we have not been able to detect any obvious diminution of Sid2p signal from one of the poles at this time. It is possible that this is due to insufficient sensitivity of our instruments or Sid2p could be rapidly replenished at the pole from cytoplasmic pools of the protein.
Although it appears from these studies that Sid2p requires microtubules and the presence of a medial ring for it to localize to the division site, it will be important in future studies to address the target(s) and/or binding partners of Sid2p at the cleavage site. Although Sid2p initially appears to localize to the medial ring, once the septum has begun to form Sid2p also localizes on either side of the developing septum. Thus, Sid2p may have targets involved in both medial ring constriction and in forming the septum. One candidate binding partner for Sid2p is Cdc15p. cdc15 mutants are capable of forming medial rings that contain actin and Cdc4p (Balasubramanian et al. 1998), but not Cdc15p (Fankhauser et al. 1995). However, Sid2p cannot localize to the medial rings that form in a cdc15 mutant. Thus, Sid2p requires Cdc15p or a Cdc15p-associated protein as a docking protein at the medial ring. The inability of cdc15 mutants, unlike mutants in other ring components, to form any deposits of septum material may be due to its inability to recruit Sid2p to the ring.
Sid2p Kinase Activity Is Essential for the Function of the Protein
The sequence of sid2 predicted that the protein may function as a kinase (Balasubramanian et al. 1998), and in this study we demonstrate that Sid2p does in fact possess in vitro kinase activity. Sid2p kinase activity is essential for the function of the protein, since a kinase-dead version of Sid2p does not display in vitro kinase activity and will not rescue the sid2-250 mutant. In fact, stronger expression of the Sid2p kinase-dead mutant has a dominant negative phenotype similar to the sid2 loss of function phenotype in either wild-type or sid2-250 cells, indicating that Sid2p-K238R may be titrating out essential regulatory factors or substrates.
Sid2p kinase activity peaks precisely when the medial ring is constricting and septum is deposited. There also appears to be some Sid2p kinase activity early in mitosis, before the anaphase peak. The presence of this activity could suggest a function for Sid2p earlier in mitosis, although the phenotype of the sid2 mutant gives no indication of this. Since total Sid2p protein levels were examined and found not to change appreciably throughout the cell cycle (data not shown), the peak in Sid2p kinase activity is presumably regulated posttranslationally. It will be important to determine in future studies if Sid2p kinase activity is regulated by phosphorylation and/or binding to regulatory subunits. Interestingly, the kinase appears to be most active during the time when it transiently accumulates at the cleavage site, suggesting that it may function to activate, by phosphorylation, the machinery at the cleavage site involved in initiating medial ring constriction and septum deposition. It will be important to determine if a single regulatory event triggers both activation of the kinase and its relocalization. It is clear that Sid2p's own kinase activity does not play a role in directing it to the cleavage site, since kinase-dead versions of Sid2p were still capable of localizing to the cleavage site.
Sid2p Functions Downstream of the Other Proteins Required to Initiate Septation
Sid2p kinase activity is diminished at the restrictive temperature in all of the mutants required for medial ring constriction and septation, suggesting that Sid2p may function at the end of the signaling cascade. A variety of other data are consistent with this hypothesis. Sid2p-GFP does not localize to both the SPB and the medial ring in sid4, cdc7, and cdc11 mutants, and it localizes to the SPB but not the medial ring in sid1, spg1, and cdc14 mutants. Thus, Sid2p does not localize properly in mutants in any of the other components of this pathway. Interestingly, although Sid2p does not localize to the medial region in the medial ring mutant cdc15, its kinase activity is unaffected, showing that localization of Sid2p to the medial ring is not necessary for the kinase to become activated. Also, Cdc7p localization is unaffected in a sid2-250 mutant, and the sid2-250 phenotype is epistatic to the Spg1p overproduction phenotype, which further supports Sid2p functioning downstream of spg1. Thus, loss of any of the putative upstream signaling molecules shuts off the cascade, leaving Sid2p in an inactive state and unable to distribute properly and presumably phosphorylate its target substrates.
The SPB as a Signaling Site for Cytokinesis
Many recent studies have begun to identify the SPB as the site of localization of many molecules that regulate cytokinesis. In S. pombe, Plo1p (Bähler et al. 1998a), Cdc7p, Spg1p (Sohrmann et al. 1998), and Sid2p (this study) all localize to the SPB and are required for initiation of medial ring constriction and septation. The reason for their localization to this site is unclear. It is possible that by virtue of their localization at the SPBs they are in a position to receive a signal that anaphase is completed and cell division can be initiated. Two proteins that are required for formation of the medial ring, the IQGAP protein Rng2p (Eng et al. 1998) and Plo1p kinase (Ohkura et al. 1995; Bähler et al., 1998), have been shown to localize to both the spindle poles and the medial ring. This theme may not be unique to fission yeast. In animal cells, a number of proteins localize to the centrosome early in mitosis and then move to the central spindle and cleavage furrow. At least two of these, the Polo and AIM-1 kinases, have been demonstrated to be required for cytokinesis (Carmena et al. 1998; Terada et al. 1998).
Is the Sid Signaling Pathway Conserved?
In Saccharomyces cerevisiae, there exists a set of genes similar to the sid group of genes in S. pombe. The S. cerevisiae genes cdc5, cdc15, tem1, and dbf2 are homologous to the S. pombe genes plo1, cdc7, spg1, and sid2, respectively. Mutations in the S. cerevisiae genes cause a block at the end of anaphase with an intact spindle and a failure in cytokinesis. Interestingly, like Sid2p, Dbf2p protein kinase activity peaks at the end of anaphase (Toyn and Johnston 1994). Recent work has suggested that the primary function of these genes may, like in S. pombe, be in cytokinesis (Jimenez et al. 1998). Like its counterpart, Plo1p, Cdc5p localizes to the SPB (Cheng et al. 1998; Shirayama et al. 1998). The other S. cerevisiae genes in this pathway have not been localized. Recently, a protein called Mob1 was identified which interacted with the Sid2p homologue, Dbf2 (Komarnitsky et al. 1998; Luca and Winey 1998). Using database searches of genome sequencing projects, we noticed that homologues of Mob1 exist in both S. pombe and humans, suggesting that this signaling pathway may be conserved in higher eukaryotes as well.
In this study we have identified the Sid2p kinase as a component of a novel signaling cascade residing at the SPB that may transmit a signal that anaphase is complete from the SPB to the cell division site, thereby causing the cell to divide. From our studies it appears that Sid2p may function downstream of the other known components of the signaling pathway. It will be important in future studies to determine how and where the other known components of this pathway function in the cascade. Although the medial ring forms early in mitosis, it does not initiate constriction until the end of anaphase after chromosome segregation. The events that take place at the ring at the end of anaphase to cause the ring to begin to constrict and the septum to form are not known. Sid2p is the first protein required for initiation of ring constriction and septation to be localized to the medial ring. The identification of Sid2p should facilitate the use of biochemical and genetic approaches to identify targets of Sid2p involved in medial ring constriction and cell division.
Acknowledgments
We are grateful to Hayes McDonald, Shelly Sazer, Pam Silver, Viesturs Simanis, and Keith Gull for providing strains and antibodies. We thank Farid Irshad for technical assistance, and Kathy Gould, Mohan Balasubramanian, and Jürg Bähler for critical reading of the manuscript.
This work was supported by National Institutes of Health (NIH) grant GM58406 to D. McCollum. C. Sparks was supported by NIH postdoctoral fellowship GM19199. M. Morphew works at the Boulder Laboratory for 3-Dimensional Fine Structure, which is supported by an NIH grant (RR0592 to J.R. McIntosh).
Footnotes
1.used in this paper: GFP, green fluorescent protein; HA, hemagglutinin; MBP, myelin basic protein; SPB, spindle pole body
References
- Bähler J., Pringle J.R. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–1370. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Steever A.B., Wheatley S., Wang Y., Pringle J.R., Gould K.L., McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast J. Cell Biol. 143 1998. 1603 1616a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A.R., Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe Yeast 14 1998. 943 951b [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., Helfman D.M., Hemmingsen S.M. A new tropomyosin essential for cytokinesis in the fission yeast S. pombe . Nature. 1992;360:84–87. doi: 10.1038/360084a0. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., Hirani B.R., Burke J.D., Gould K.L. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J. Cell Biol. 1994;125:1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M.K., McCollum D., Gould K.L. Cytokinesis in fission yeast Schizosaccharomyces pombe . Methods Enzymol. 1997;283:494–506. doi: 10.1016/s0076-6879(97)83039-x. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., McCollum D., Chang L., Wong K.C., Naqvi N.I., He X., Sazer S., Gould K.L. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N., Muriel W.J., Carr A.M. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe . Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Basi G., Schmid E., Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D.J., Salmon E.D., McMillan J.N., Yeh E., Pringle J.R. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Riparbelli M.G., Minestrini G., Tavares A.M., Adams R., Callaini G., Glover D.M. Drosophila polo kinase is required for cytokinesis. J. Cell Biol. 1998;143:659–671. doi: 10.1083/jcb.143.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Nurse P. How fission yeast fission in the middle. Cell. 1996;84:191–194. doi: 10.1016/s0092-8674(00)80973-3. [DOI] [PubMed] [Google Scholar]
- Chang F., Wollard A., Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Cheng L., Hunke L., Hardy C.F.J. Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p. Mol. Cell. Biol. 1998;18:7360–7370. doi: 10.1128/mcb.18.12.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R.A., Griffiths D.J., Sheldrick K.S., Randall R.E., Hagan I.M., Carr A.M. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe . Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- Ding R., West R.R., Morphew D.M., Oakley B.R., McIntosh J.R. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng K., Naqvi N.I., Wong K.C., Balasubramanian M.K. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr. Biol. 1998;8:611–621. doi: 10.1016/s0960-9822(98)70248-9. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Simanis V. The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:3011–3019. doi: 10.1002/j.1460-2075.1994.tb06600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Reymond A., Cerutti L., Utzig S., Hoffmann K., Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 1995;82:435–444. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- Furge K.A., Wong K., Armstrong J., Balasubramanian M., Albright C.F. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol. 1998;8:947–954. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- Gould K., Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. Evidence for cell cycle–specific, spindle pole body–mediated, nuclear positioning in the fission yeast Schizosaccharomyces pombe . J. Cell Sci. 1997;110:1851–1866. doi: 10.1242/jcs.110.16.1851. [DOI] [PubMed] [Google Scholar]
- Hagan I.M. The fission yeast microtubule cytoskeleton. J. Cell Sci. 1998;111:1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- He X., Patterson T.E., Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C.S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli . Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Jimenez J., Cid V.J., Cenamor R., Yuste M., Molero G., Nombela C., Sanchez M. Morphogenesis beyond cytokinetic arrest in Saccharomyces cerevisiae . J. Cell Biol. 1998;143:1617–1634. doi: 10.1083/jcb.143.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney J.B., Boeke J.D. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe . Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama C., Sugimoto A., Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe . J. Cell Biol. 1997;137:1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky S.I., Chiang Y.C., Luca F.C., Chen J., Toyn J.H., Winey M., Johnston L.H., Denis C.L. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupold U. Methods in Cell Physiology Academic Press NewYork. 41970. 169 177 [Google Scholar]
- Luca F.C., Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks J., Fankhauser C., Simanis V. Genetic interactions in the control of septation in Schizosaccharomyces pombe . J. Cell Sci. 1992;101:801–808. doi: 10.1242/jcs.101.4.801. [DOI] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- McCollum D., Feoktistova A., Morphew M., Balasubramanian M., Gould K.L. The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast, Schizosaccharomyces pombe . Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux P., Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe . Mol. Gen. Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Ohkura H., Hagan I.M., Glover D.M. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- Pichova A., Kohlwein S.D., Yamamoto M. New arrays of cytoplasmic microtubules in the fission yeast Schizosaccharomyces pombe . Protoplasma. 1995;188:252–257. [Google Scholar]
- Prentice H.L. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport R. Establishment of the mechanism of cytokinesis in animal cells. Int. Rev. Cytol. 1986;105:245–281. doi: 10.1016/s0074-7696(08)61065-7. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular CloningA Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Satterwhite L.L., Pollard T.D. Cytokinesis. Curr. Opin. Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- Schmidt S., Sohrmann M., Hofmann K., Woollard A., Simanis V. The Spg1p GTPase is an essential dosage-dependent inducer of septum formation in Schizosaccharomyces pombe . Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Zachariae W., Ciosk R., Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae . EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M., Fankhauser C., Brodbeck C., Simanis V. The dmf/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Sohrmann M., Schmidt S., Hagan I., Simanis V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 1998;12:84–94. doi: 10.1101/gad.12.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y., Tatsuka M., Suzuki F., Yasuda Y., Fujita S., Otsu M. AIM-1a mammalian midbody-associated protein required for cytokinesis. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn J.H., Johnston L.H. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley S.P., Wang Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]