Abstract
In yeast, mitochondrial division and fusion are highly regulated during growth, mating and sporulation, yet the mechanisms controlling these activities are unknown. Using a novel screen, we isolated mutants in which mitochondria lose their normal structure, and instead form a large network of interconnected tubules. These mutants, which appear defective in mitochondrial division, all carried mutations in DNM1, a dynamin-related protein that localizes to mitochondria. We also isolated mutants containing numerous mitochondrial fragments. These mutants were defective in FZO1, a gene previously shown to be required for mitochondrial fusion. Surprisingly, we found that in dnm1 fzo1 double mutants, normal mitochondrial shape is restored. Induction of Dnm1p expression in dnm1 fzo1 cells caused rapid fragmentation of mitochondria. We propose that dnm1 mutants are defective in the mitochondrial division, an activity antagonistic to fusion. Our results thus suggest that mitochondrial shape is normally controlled by a balance between division and fusion which requires Dnm1p and Fzo1p, respectively.
Keywords: mitochondrial division, mitochondrial fusion, dynamin, GTPase, yeast
Mitochondria undergo regulated fusion and division in many cell types ( Bereiter-Hahn and Voth 1994; Kawano et al. 1995), which appear to play key roles in establishing and maintaining mitochondrial shape ( Tyler 1992). In the yeast S. cerevisiae, mitochondria are elongated, tubule-shaped organelles that are very dynamic during growth, mating, and sporulation. Mitochondria constitutively divide and fuse during cell growth ( Nunnari et al. 1997), but change their number depending on growth conditions ( Stevens 1977). During mating, mitochondria fuse immediately after cell fusion, mixing their contents, including mitochodrial DNA (mtDNA) and matrix proteins ( Nunnari et al. 1997; Okamoto et al. 1998). When diploids sporulate, mitochondria are dramatically reorganized moving into the four spores and surrounding each haploid nucleus ( Miyakawa et al. 1984). The yeast homologue of fuzzy onions ( Hales and Fuller 1997), FZO1 was recently identified and shown to play an important role in mitochondrial fusion ( Hermann et al. 1998; Rapaport et al. 1998). However, the mechanisms that control mitochondrial division are unknown.
Materials and Methods
Strain Construction
Strain YHS2, which expresses red-shifted GFP (F64L, T65C, and I167T) fused to the Cox4p presequence (residues 1–21) under the ADH1 promoter (ADH1-COX4-GFP), was constructed as follows. First, pHS1 was constructed by replacing wild-type GFP in pOK29, a HIS3-CEN plasmid which carries ADH1-COX4-GFP (Kerscher, O., unpublished information), with a NcoI-BamHI fragment carrying red-shifted GFP from pQBI25 (Quantum Biotechnologies). The EcoRV-BamHI fragment from pHS1 was inserted into pDH9, which carries 5′ and 3′ untranslated regions of MFA2 (a gift from S. Michaelis), forming pHS2. To integrate ADH1-COX4-GFP at chromosomal MFA2, a XhoI-SmaI fragment carrying 5′-MFA2-ADH1-COX4-GFP-3′-MFA2 from pHS2 was transformed into strain SM1235, which carries mfa2::URA3 ( Michaelis and Herskowitz 1988). Strain YHS2, which contains MATα mfa2::ADH1-COX4-GFP, was selected on 5-fluoro-orotic acid medium ( Adams et al. 1997). MAT a mfa2::ADH1-COX4-GFP strain YHS1 was obtained by crossing YHS2 to SM1227 ( Michaelis and Herskowitz 1988). Strain 1002 (MATa, his3, trp1, ura3, mfa2::ADH1-COX4-GFP) was constructed by crossing YHS1 to BY4731 ( Brachmann et al. 1998).
Mutant Isolation
YHS2 was mutagenized with 3% ethane methylsulfonate to ∼30% survival ( Adams et al. 1997). Mutagenized cells were suspended at ∼9 × 105 cells/ml on coverslips and observed using an inverted microscope and the HIQ GFP 41014 filter set (Chroma). Mutants were isolated using micropipettes (10 μm diameter; World Precision Instruments), transferred to a drop of SD on the same coverslip, and then to YPD plates. Micropipettes were handled by an Eppendorf micromanipulator 5171.
Crosses to wild-type strain 1002 showed that all class I, II, and III mutations were recessive and caused by a defect in a single gene. Complementation tests revealed that all eight recessive class IV mutants were defective in the same gene. Crosses between class IV mutants and TRP1 strain 194 (a gift from E. Schweizer) or dnm1Δ strain JSY1361 ( Otsuga et al. 1998) showed the class IV mutation was centromere linked, located on chromosome XII, and allelic to dnm1. DNM1-containing plasmid, pRU1-DNM1 ( Otsuga et al. 1998), rescued the mitochondrial phenotype of all recessive class IV mutants. All dominant and semidominant class IV mutations also segregated as alleles of dnm1.
Gene Disruption
Complete disruptions of the DNM1 and FZO1 were constructed by PCR-mediated gene replacement as described ( Lorenz et al. 1995) into strains BY4733 and BY4744 ( Brachmann et al. 1998). For dnm1Δ, we used HIS3 plasmid pRS303 ( Sikorski and Hieter 1989) and for fzo1Δ we used kanMX4 plasmid pRS400 ( Brachmann et al. 1998). MATa dnm1Δ fzo1Δ strain YHS27 and MATα dnm1Δ fzo1Δ strain YHS23 were constructed by crossing MATa dnm1Δ strain YHS19 to MATα fzo1Δ strain YHS22. Mitochondria in the disruption strains were visualized using pHS12, a CEN-LEU2 plasmid containing ADH1-COX4-GFP. pHS12 was created by inserting the XhoI-NotI fragment from pHS1 into pRS315 ( Sikorski and Hieter 1989).
Plasmid Construction
The DNM1 gene with a NotI site immediately preceding its termination codon was PCR amplified from yeast genomic DNA and subcloned into pAA3, a CEN-LEU2 plasmid which contains the HA epitope with a NotI site at its NH2 terminus (Aiken, A., unpublished data), forming pDNM1-HA (pHS14). DNM1-GFP plasmid pHS20 was constructed as described above except that pAA1, a CEN-LEU2 plasmid which contains GFP with a NotI site at its NH2 terminus (Aiken, A., unpublished data), was used instead of pAA3. To form pHS15, DNM1-HA coding sequences were PCR amplified from pHS14 with 50 bp of flanking sequences homologous to the GAL1-URA3 promoter in pRS314GU ( Nigro et al. 1992). The DNM1-HA fragment and linearized pRS314GU were cotransformed into yeast and pGAL1-DNM1-HA (pHS15) was formed by homologous recombination ( Oldenburg et al. 1997). pGAL1-DNM1-GFP (pHS40) was constructed as described for pHS15 except that pHS20 was used instead of pHS14.
Quantitation of Dnm1p-GFP Localization
dnm1Δ fzo1Δ cells carrying pGAL1-DNM1-GFP were incubated in galactose media for 1–2 h, stained with MitoTracker Red CMXRos (Molecular Probes). 12 cells were examined by fluorescence microscopy and the mitochondrially associated Dnm1p-GFP dots (82 total) were assigned to one of two locations: (a) the end of a tubule (50 dots), or (b) the side of a tubule (32 dots). The end of a tubule was defined as when the center of a Dnm1p-GFP dot was located within 0.15 μm from the end. The average length of the mitochondrial tubules was estimated to be 2.7 ± 1.9 μm and the diameter ∼0.3 μm (n = 44). We calculate that the side of the tubule represents 89% of the mitochondrial surface area and the remainder (11%) represents the end.
Results and Discussion
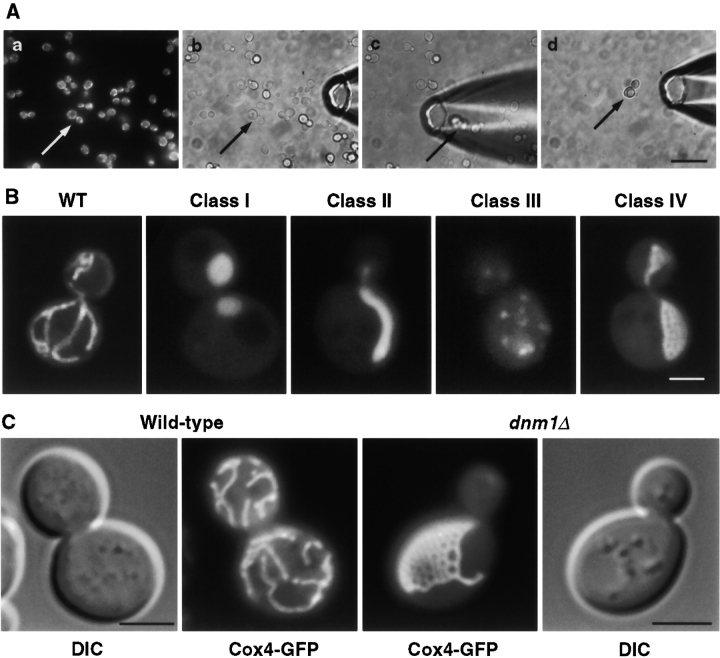
We screened for yeast mutants defective in mitochondrial shape using a novel strategy in which mitochondria are visualized by the green fluorescent protein (GFP) and mutants were isolated by micromanipulation. GFP was fused to the presequence (residues 1–21) of mitochondrial cytochrome oxidase subunit IV (COX4; Pon and Schatz 1991). When expressed in yeast, COX4-GFP targets the mitochondrial matrix, and mitochondria were visible by fluorescence microscopy. We integrated the COX4-GFP gene at the nonessential MFA2 locus ( Michaelis and Herskowitz 1988), which made fluorescence intensity uniform among cells and enabled efficient screening. After mutagenesis, individual cells with abnormal mitochondrial shape were hand-isolated using micropipettes ( Fig. 1 A). This screening procedure allowed us to isolate individual mutant cells with interesting mitochondrial phenotypes from a large total population of cells.
Figure 1.
Mutants defective in mitochondrial shape. (A) Screen to isolate mutants defective in mitochondrial morphology. (a) Mitochondria were visualized by COX4-GFP. (b–d) Individual mutants were isolated using a micropipette. The arrow indicates a potential mutant cell. (B) Wild-type YHS2 and a representative from each class of mutants were grown in YPD to log phase and then stained with 1 μg/ml 3,3′dihexyloxacarbocyanine (DiOC6) (Molecular Probes) to enhance mitochondrial fluorescence. Confocal images of cells are shown. (C) dnm1Δ cells contain an interconnected network of mitochondria. Wild-type (BY4733) and isogenic dnm1Δ cells expressing COX4-GFP (pHS12), were grown in YPGal to log phase. Fluorescence (COX4-GFP) and differential interference contrast (DIC) images are shown. Bars: (A) 10 μm; (B) 2 μm; (C) 3 μm.
Of ∼72,000 cells screened, we isolated 20 mutants, which were classified into four categories ( Fig. 1 B). Class I mutants (two isolates) contained one or two large, spherical mitochondria instead of the normal tubules seen in wild-type cells. Genetic crosses showed that both carried mdm10 mutants ( Sogo and Yaffe 1994). The single class II mutant contained one or two oblong mitochondria collapsed to one side of the cell and was found to be defective in SLM1. slm1 was previously identified as an mmm1 synthetic-lethal mutant ( Burgess et al. 1994; Burgess et al., manuscript in preparation). Class III mutants (three isolates) contained numerous mitochondrial fragments and were shown to carry fzo1 mutations. FZO1 encodes a GTPase anchored in the mitochondrial outer membrane that is required for mitochondrial fusion ( Hermann et al. 1998; Rapaport et al. 1998). Class IV mutants (14 isolates) exhibited a novel phenotype consisting of an interconnected network of mitochondrial tubules. In contrast to wild-type, which have 5–10 separate mitochondria per cell, class IV mutants appear to contain a single organelle. Because of their unique networked mitochondrial shape, these mutants were examined further.
Genetic crosses showed that our 14 class IV mutants comprised 8 recessive, 5 dominant and 1 semi-dominant mutations. Mapping studies showed that all 14 mutations were centromere linked (1.1 cM) and located on chromosome XII. We noted that DNM1 ( Gammie et al. 1995), a gene related to dynamin GTPase ( Obar et al. 1990), maps to chromosome XII near the centromere and is required for mitochondrial shape ( Otsuga et al. 1998). Using a dnm1Δ strain and a plasmid containing DNM1 (kindly provided by J. Shaw), we found that all 14 class IV mutants carried dnm1 alleles. These results were unexpected since mitochondrial shape in our mutants was strikingly different from previously seen in dnm1 mutants, where mitochondria collapse to one side of the cell and form a single tubule ( Otsuga et al. 1998).
A complete disruption of DNM1 coding sequences was constructed, and examined for mitochondrial shape ( Fig. 1 C). ∼90% of dnm1Δ cells showed a single highly branched mitochondrial network. ∼10% of dnm1Δ cells displayed a single mitochondrial tubule localized to one side of the cell, similar to that seen earlier ( Otsuga et al. 1998). The mitochondrial shape was not dramatically altered by growth conditions (not shown). Mitochondria in dnm1 mutants were efficiently segregated during cell division ( Fig. 1 B and 2). Small daughter buds often contained a single mitochondrial tubule without branches, while larger buds had small networks. Most mitochondria were continuous from mother cells to buds, and separate mitochondria were only seen after the two cells separated. Our results strongly suggest that dnm1 mutants are defective in mitochondrial division. We speculate that mitochondria in dnm1 mutants may be divided indirectly, perhaps by cytokinesis. The yeast cell division machinery is clearly robust enough for the job, since nuclei are efficiently severed by cytokinesis in S. pombe cut mutants ( Uzawa et al. 1990).
In yeast, mitochondria are very dynamic, fusing or dividing on average every two minutes ( Nunnari et al. 1997). Thus there appears to be an equilibrium between fusion and division. Supporting this idea, when FZO1, a gene required for mitochondrial fusion, is defective, mitochondria fragment due to continued fission of the organelle ( Hermann et al. 1998; Rapaport et al. 1998). We hypothesized that if mitochondrial division were blocked, cells would have fewer (larger) organelles. Our working model, based on the morphology of dnm1 mutants, is that Dnm1p is required for mitochondrial division. To test this hypothesis, we constructed double mutants containing both dnm1Δ and fzo1Δ by genetic crosses. We found that normal mitochondrial shape was restored ( Fig. 2). ∼85% of double mutants contained multiply-branched, tubular mitochondria very similar to those seen in wild-type cells (Table ). This was in marked contrast to dnm1Δ mutants, which usually had a single organelle, and fzo1Δ mutants, with numerous mitochondrial fragments ( Fig. 2; Hermann et al. 1998; Rapaport et al. 1998). Mitochondria in dnm1Δ fzo1Δ cells were not always completely normal; the tubules tended to be longer and more curved than in wild-type cells, and occasionally formed bundles. Nonetheless, our observations suggest that excess mitochondrial division in fzo1Δ cells is suppressed by inactivating DNM1, and that excess mitochondrial fusion in dnm1Δ cells is rescued by fzo1Δ. We propose that division, which requires Dnm1p, and fusion, controlled by Fzo1p, have antagonistic effects on mitochondrial shape and number. Our results also suggest that mitochondrial tubule formation occurs by a mechanism independent of fusion and division.
Figure 2.
dnm1Δ fzo1Δ cells have normal-shaped mitochondria. Wild-type (BY4733) and isogenic deletion strains (dnm1Δ, fzo1Δ, and dnm1Δ fzo1Δ) expressing COX4-GFP (pHS12), were grown in YPGal to log phase and examined by fluorescence microscopy. Bars, 2 μm.
Table 1.
Mitochondrial Morphology in dnm1ΔfzolΔ Cells Created by Consecutive Gene Disruption or Genetic Cross
| Mitochondrialmorphology | dnm1ΔfzolΔ Cells created byconsecutive gene disruption | dnm1ΔfzolΔ Cellscreated by cross | |
|---|---|---|---|
| fzolΔ→dnm1Δ | dnm1Δ→fzolΔ | ||
| % | % | % | |
| Tubules | 50.0 ± 7.5 | 60.1 ± 4.7 | 84.7 ± 3.4 |
| Networks | 0.3 ± 0.7 | 30.6 ± 3.6 | 0.3 ± 0.6 |
| Fragments | 39.9 ± 5.2 | 0 ± 0 | 0 ± 0 |
| Other | 9.8 ± 3.3 | 9.3 ± 3.1 | 14.9 ± 2.9 |
Interestingly, mitochondrial shape and number in dnm1Δ fzo1Δ cells was dependent upon the order of gene disruption. When cells were first disrupted for FZO1 and subsequently for DNM1 (Table , fzo1Δ→dnm1Δ), ∼40% of cells carried mitochondrial fragments similar to those seen in fzo1Δ single mutants. In contrast, when cells were first disrupted for DNM1 and then for FZO1 (Table , dnm1Δ→fzo1Δ), ∼30% of cells displayed a mitochondrial network like that seen in dnm1Δ cells. Our results indicate that the mitochondrial networks found in dnm1Δ mutants persist in the absence of fusion activity, and fragments formed in the fzo1Δ mutant persist in the absence of fission activity. We also found tubular mitochondria in many of the double mutant cells formed by consecutive gene disruption (∼50% for fzo1Δ→dnm1Δ; ∼60% for dnm1Δ→ fzo1Δ). These results further indicate that mitochondrial tubules form in the absence of division and fusion. It is not clear why dnm1Δ fzo1Δ double mutants generated by crossing a dnm1Δ cell to a fzo1Δ cell contained mostly (>80%) tubular mitochondria and essentially no mitochondrial networks or fragments (Table ). During germination and growth of a dnm1Δ fzo1Δ spore, it is possible that cells are simultaneously depleted of Dnm1p and Fzo1p, leading to the formation of tubules, but not networks or fragments.
To further test the role of Dnm1p in division, we induced Dnm1p expression in dnm1Δ fzo1Δ cells and observed its effect on mitochondria. Dnm1p was fused to the HA epitope (Dnm1p-HA) ( Field et al. 1988) and expressed under the galactose-inducible GAL1 promoter ( Nigro et al. 1992). Our pGAL1-DNM1-HA rescued the dnm1Δ phenotype on galactose medium (not shown). When dnm1Δ fzo1Δ cells containing pGAL1-DNM1-HA were grown in the absence of galactose, no Dnm1p-HA was detected ( Fig. 3 B) and ∼70% of cells displayed the tubular mitochondria typical of dnm1Δ fzo1Δ mutants ( Fig. 3 A). Upon transfer to galactose medium, Dnm1p-HA levels gradually increased, while the level of hexokinase, a control protein, remained constant ( Fig. 3 B). Concomitant with the accumulation of Dnm1p-HA, mitochondrial shape changed dramatically ( Fig. 3 A). The number of cells with tubular mitochondria decreased, and those with fragmented mitochondria increased. By 5 h, ∼65% of the cells contained completely fragmented mitochondria. At intermediate times (2 h) after inducing Dnm1p-HA, cells contained partially fragmented tubules, and many mitochondrial tubules were adjacent to small fragments. Our results clearly show that the division of mitochondria in dnm1Δ fzo1Δ cells coincides with the expression of Dnm1p.
Figure 3.
Expression of Dnm1p causes fragmentation of mitochondria in dnm1Δ fzo1Δ cells. dnm1Δ fzo1Δ cells carrying pCOX4-GFP (pHS12) and pGAL1-DNM1-HA (pHS15) were pregrown in raffinose medium, centrifuged and resuspended to an OD600 of 0.2 in galactose medium (SGS) to induce Dnm1p-HA expression. Cells were examined for mitochondrial shape (n = 100) or used to prepare total protein at the indicated timepoints. (A) Mitochondrial shape was classified into the following four groups: tubules (○), partially fragmented tubules (□), fragments (▴), and other (⋄). (B) Total protein was extracted as described ( Yaffe and Schatz 1984) and subjected to Western blot analysis using antibodies to the HA epitope ( Field et al. 1988), or hexokinase (Davis, A., unpublished data) followed by chemiluminescence (Pierce). Relative amounts of Dnm1p-HA (+) were quantitated and plotted in A.
Further supporting a role for Dnm1p in mitochondrial fission, we found that the Dnm1 protein was preferentially localized to sites of mitochondrial division. We constructed a fusion between Dnm1p and the green fluorescent protein (GFP). Consistent with previous results ( Otsuga et al. 1998), we found that much of Dnm1p-GFP was associated with mitochondria in punctate structures ( Fig. 4 A). In cells that were constitutively expressing Dnm1p-GFP, it was difficult to determine the precise location of Dnm1p because of the complex morphology of the mitochondria and the large number of Dnm1p-GFP dots. To simplify our analyses, we induced the expression of Dnm1p-GFP in the dnm1Δ fzo1Δ mutant and examined cells at early times after induction. We found a tight correlation between the appearance of Dnm1p-GFP and fragmentation of mitochondria. Two representative cells are shown in Fig. 4B and Fig. C; both cells contained two Dnm1p-GFP dots, one of which was located at the end of a tubule, the other appeared to be reside near a constricted region of the mitochondrion. After analysis of additional cells, we found that Dnm1p-GFP was localized to ends of mitochondrial fragments much more frequently (>60%) than predicted if Dnm1p-GFP was randomly distributed on mitochondria (∼11%). These results suggest that Dnm1p acts at the site of mitochondrial fission. We note that Dnm1p-GFP is not exclusively found at the ends of mitochondria. We surmise that Dnm1p on the sides of the tubules may mark future sites of division, or represent Dnm1p-containing complexes that have diffused away from the end of the tubule. More definitive experiments (e.g., time-lapse videomicroscopy) to determine the role of Dnm1p in fission are in progress.
Figure 4.
A Dnm1p-GFP preferentially localizes to site of mitochondrial division. (A) dnm1Δ strain YHS19 was transformed with pHS20 expressing Dnm1p-GFP. Cells were grown in SGal medium, labeled with 0.1 μM MitoTracker Red CMXRos (Molecular Probes) and then examined under the fluorescence microscope. Merged images taken in the red (Mitotracker) and green (GFP) channels are shown. (B and C) dnm1Δ fzo1Δ diploid cells carrying pGAL1-DNM1-GFP were pregrown in YPglycerol/ethanol medium, centrifuged and grown in galactose medium (SGS) for 1–2 h. Cells were then stained with MitoTracker and examined for mitochondrial localization of Dnm1p-GFP. Two representative cells are shown. Bar, 3 μm.
The relatively normal mitochondria seen in dnm1Δ fzo1Δ mutants could be explained by a restoration of fusion activity; for example, if Dnm1p were an inhibitor of mitochondrial fusion. To test this possibility, we monitored mitochondrial fusion during mating ( Nunnari et al. 1997; Okamoto et al. 1998). Mitochondria were visualized in one parent, by the galactose-induced expression of a matrix-targeted GFP (CS1-GFP) on plasmid pCLbGFP ( Okamoto et al. 1998). MATa cells containing pCLbGFP were pregrown in galactose medium to induce CS1-GFP expression, then transferred to glucose to inhibit further synthesis. MATa cells were then mixed with MATα cells, which did not carry pCLbGFP, and allowed to mate on glucose medium. Mitochondria were visualized in zygotes using MitoTracker. If mitochondrial fusion occurred, GFP and MitoTracker fluorescence would completely overlap, because the matrix-localized CS1-GFP from the MATa mitochondria diffused into the mitochondrial matrix of the MATα cell. If no fusion occurred, GFP-labeled mitochondria would be seen in only one half of the zygote.
Zygotes formed by two wild-type cells, or two dnm1Δ mutants, exhibited efficient mitochondrial fusion, with GFP fluorescence and MitoTracker overlapping in all mitochondria ( Fig. 4 A). In contrast, fusion was defective in fzo1Δ mutants, consistent with previous observations ( Hermann et al. 1998). Mitochondrial fragments tended to aggregate in fzo1Δ cells and individual fragments were difficult to distinguish. Nonetheless, in matings between two fzo1Δ cells, MitoTracker showed clusters of fragmented mitochondria in the zygote and diploid bud, but we detected GFP fluorescence in only half of the mitochondrial clusters. Like fzo1Δ mutants, dnm1Δ fzo1Δ double mutants failed to fuse their mitochondria. Although mitochondria in dnm1Δ fzo1Δ/dnm1Δ fzo1Δ diploid cells had normal shape, only half of the organelles contained GFP. Our results indicate that dnm1Δ fzo1Δ cells are defective in mitochondrial fusion.
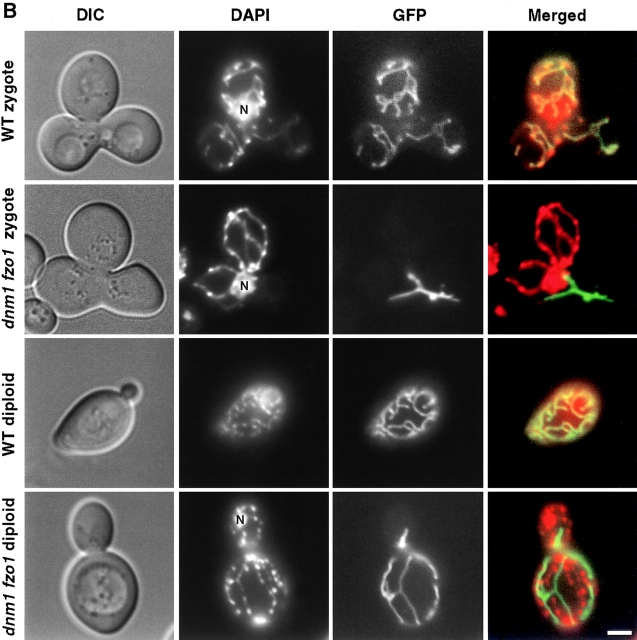
To eliminate the possibility that low, basal levels of fusion occur in dnm1Δ fzo1Δ cells, we used a more sensitive fusion assay using the matrix markers, CS1-GFP and mitochondrial DNA (mtDNA; Okamoto et al. 1998). 4′,6-diamidino-2-phenylindole (DAPI) stained mtDNA was a more stable probe compared with MitoTracker, allowing us to examine cells for longer times following the initial mating event. MATa cells, which lacked mtDNA and carried pCLbGFP were mated to MATα cells, which contained mtDNA, but not the plasmid ( Fig. 5 B). The DAPI and GFP fluorescence overlapped in all the mitochondrial tubules in wild-type zygotes (52 zygotes examined). When 500 dnm1Δ fzo1Δ zygotes were examined, we found no overlap between DAPI and GFP. The fusion activity in dnm1Δ fzo1Δ mutants is therefore at least 500-fold less than that in wild-type cells. Even after the zygotes were allowed to grow and divide, we found no fusion in the mutant cells ( Fig. 5 B). When 100 dnm1Δ fzo1Δ/dnm1Δ fzo1Δ diploid cells were examined 24 h after mating, none contained an overlap between GFP and DAPI, whereas a complete GFP and DAPI overlap was seen in 43 wild-type diploids. dnm1Δ fzo1Δ cells clearly lack significant mitochondrial fusion activity. Our results above also suggest that dnm1Δ fzo1Δ cells lack fission activity. We therefore propose that in cells lacking Fzo1p and Dnm1p, mitochondrial tubule formation occurs by a mechanism independent of fusion and division, such as growth from the ends of preexisting organelles.
Figure 5.
dnm1Δ fzo1Δ cells are defective in mitochondrial fusion. (A) MATa cells that express CS1-GFP from the GAL1 promoter of pCLbGFP were grown in galactose-containing medium (SGS). MATα cells lacking the plasmid were grown in glucose medium (YPD). MATa and α cells carrying the indicated genotypes were mated on YPD for 3.5 h, and then stained with 0.1 μM MitoTracker. The distribution of MitoTracker and GFP in representative zygotes containing a medial bud (asterisks) are shown. Arrows indicate three clusters of mitochondria lacking CS1-GFP in fzo1Δ zygotes. (B) Wild-type and dnm1Δ fzo1Δ MATa cells were treated with ethidium bromide to induce loss of mtDNA ( Fox et al. 1991), transformed with pCLbGFP, and then grown in galactose-containing medium (SGS). Wild-type and dnm1Δ fzo1Δ MATα cells containing mtDNA, but lacking the plasmid, were grown in YPD. Cells were mated on YPD and stained with 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes). A representative zygote formed by mating between wild-type cells or between dnm1Δ fzo1Δ mutants is shown. Fluorescence (GFP or DAPI), differential interference contrast (DIC) and merged images are shown. Mating mixtures were also incubated for an additional 24 h at 30°C on YPD, and representative diploids are shown. N indicates nuclear DNA staining. Bar, 3 μm.

Dynamin has been proposed to work as a mechanochemical enzyme that ‘pinches off’ plasma membrane invaginations, forming intracellular vesicles ( Takei et al. 1995). Supporting this idea, dynamin self-assembles into spiral-like structures ( Hinshaw and Schmid 1995) which can sever artificial membranes in vitro ( Sweitzer and Hinshaw 1998). It is also possible that dynamin plays a regulatory, instead of an enzymatic, function in membrane scission ( Sever et al. 1999). Future studies are clearly needed to determine the precise mechanism that Dnm1p plays in mitochondrial division.
Acknowledgments
We especially thank J. Shaw for generous gifts of strains and plasmids. We also thank J. Holder, O. Kerscher, S. Michaelis, J. Boeke, A. Aiken, A. Davis, R. Butow, K. Okamoto, D. Murphy, R. Bustos, C. Machamer, and T. Kai for reagents, equipment and technical advice. We thank C. Machamer, K. Wilson, and the members of the Jensen lab for valuable comments on the manuscript.
This work was supported by a PHS grant (R01-GM54021) to R.E. Jensen and a JSPS fellowship to H. Sesaki.
Footnotes
Abbreviations used in this paper: GFP, green fluorescent protein; mtDNA, mitochondrial DNA.
References
- Adams A., Gottschling D., Kaiser C., Stearns T. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Plainview, NY : 1997. [Google Scholar]
- Bereiter-Hahn J., Voth M. Dynamics of mitochondria in living cellsshape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994;27:198–219 . doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains from Saccharomyces cerevisiae 288Ca useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132 . doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Burgess S.M., Delannoy M., Jensen R.E. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol. 1994;126:1375–1391 . doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I.A., Lerner R.A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 1988;8:2159–2165 . doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T.D., Folley L.S., Mulero J.J., McMullin T.W., Thorsness P.E., Hedin L.O., Costanzo M.C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165 . doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Gammie A.E., Kurihara L.J., Vallee R.B., Rose M.D. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J. Cell Biol. 1995;130:553–566 . doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales K.G., Fuller M.T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129 . doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., Shaw J.M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–373 . doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J.E., Schmid S.L. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192 . doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Kawano S., Takano H., Kuroiwa T. Sexuality of mitochondriafusion, recombination, and plasmids. Int. Rev. Cytol. 1995;161:49–110 . doi: 10.1016/s0074-7696(08)62496-1. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C., Muir R.S., Lim E., McElver J., Weber S.C., Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae . Gene. 1995;158:113–117 . doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- Michaelis S., Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol. Cell. Biol. 1988;8:1309–1318 . doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa I., Aoi H., Sando N., Kuroiwa T. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae . J. Cell Sci. 1984;66:21–38 . doi: 10.1242/jcs.66.1.21. [DOI] [PubMed] [Google Scholar]
- Nigro J.M., Sikorski R., Reed S.I., Vogelstein B. Human p53 and CDC2Hs genes combine to inhibit the proliferation of Saccharomyces cerevisiae . Mol. Cell. Biol. 1992;12:1357–1365 . doi: 10.1128/mcb.12.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Marshall W.F., Straight A., Murray A., Sedat J.W., Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 1997;8:1233–1242 . doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar R.A., Collins C.A., Hammarback J.A., Shpetner H.S., Vallee R.B. Molecular cloning of the microtubule-associated mechanochemical enzyme dynamin reveals homology with a new family of GTP-binding proteins. Nature. 1990;347:256–261 . doi: 10.1038/347256a0. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Perlman P.S., Butow R.A. The sorting of mitochondrial DNA and mitochondrial proteins in zygotespreferential transmission of mitochondrial DNA to the medial bud. J. Cell Biol. 1998;142:613–623 . doi: 10.1083/jcb.142.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg K.R., Vo K.T., Michaelis S., Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452 . doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D., Keegan B.R., Brisch E., Thatcher J.W., Hermann G.J., Bleazard W., Shaw J.M. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998;143:333–349 . doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon L., Schatz G. Biogenesis of yeast mitochondria. In: Broach J.R., Pringle J.R., Jones E.W., editors. The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press; New York : 1991. pp. 333–406. [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae . J. Biol. Chem. 1998;273:20150–20155 . doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Sever S., Muhlberg A.B., Schmid S.L. Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486 . doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- Sikorski R., Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–28 . doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo L.F., Yaffe M.P. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 1994;126:1361–1373 . doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B.J. Variation in number and volume of the mitochondria in yeast according to growth conditions. A study based on serial sectioning and computer graphics reconstitution. Biol. Cell. 1977;28:37–56 . [Google Scholar]
- Sweitzer S.M., Hinshaw J.E. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029 . doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Takei K., McPherson P.S., Schmid S.L., De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–190 . doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Tyler D. The mitochondrion. VCH Publishers; New York : 1992. [Google Scholar]
- Uzawa S., Samejima I., Hirano T., Tanaka K., Yanagida M. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell. 1990;62:913–925 . doi: 10.1016/0092-8674(90)90266-h. [DOI] [PubMed] [Google Scholar]
- Yaffe M.P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]