Abstract
Tom40 is an essential component of the preprotein translocase of the mitochondrial outer membrane (TOM complex) in which it constitutes the core element of the protein conducting pore. We have investigated the biogenesis of Tom40. Tom40 is inserted into the outer membrane by the TOM complex. Initially, Tom40 is bound as a monomer at the mitochondrial surface. The import receptor Tom20 is involved in this initial step; it stimulates both binding and efficient insertion of the Tom40 precursor. This step is followed by the formation of a further intermediate at which the Tom40 precursor is partially inserted into the outer membrane. Finally, Tom40 is integrated into preexisting TOM complexes. Efficient import appears to require the Tom40 precursor to be in a partially folded conformation. Neither the NH2 nor the COOH termini are necessary to target Tom40 to the outer membrane. However, the NH2-terminal segment is required for Tom40 to become assembled into the TOM complex. A model for the biogenesis of Tom40 is presented.
Keywords: mitochondrial protein import, TOM complex, Tom40, protein insertion, unfolding
Transport of proteins into and across the two mitochondrial membranes is achieved through the concerted action of translocation machineries: the TOM complex in the outer membrane, and one of the two TIM complexes in the inner membrane (Glick and Schatz 1991; Pfanner et al. 1992; Lill and Neupert 1996; Schatz and Dobberstein 1996). Targeting and initial translocation of most preproteins that are destined to the mitochondrial matrix are dependent on an NH2-terminal cleavable presequence (Haucke and Schatz 1997; Neupert 1997). In contrast, proteins of the outer membrane, and a number of proteins of the inner membrane and the intermembrane space, contain noncleavable targeting signals (Shore et al. 1995; Stuart and Neupert 1996; Neupert 1997). Currently, the nature of most of these latter signals is obscure.
The TOM complex contains import receptors for the initial recognition of preproteins (Tom20, Tom22, and Tom70) and membrane-embedded components that form the general import pore which facilitates the translocation of preprotein across the outer membrane (Tom40, Tom5, Tom6, and Tom7). Tom40, a protein essential for viability of yeast and Neurospora crassa cells, was found to be the most abundant component of the TOM complex (Dekker et al. 1998; Künkele et al. 1998a) and the core element of the preprotein-conducting pore (Hill et al. 1998; Künkele et al. 1998b). The protein forms oligomers with dimers as the basic structure and it was found to interact with polypeptide chains in transit (Vestweber et al. 1989; Kiebler et al. 1990; Rapaport et al. 1997, Rapaport et al. 1998b). During preprotein translocation, the Tom40 oligomer undergoes conformational changes that affect both the structure of the Tom40 dimer and its interaction with other constituents of the TOM complex (Rapaport et al. 1998b). Tom40 is predicted to traverse the outer membrane as a series of 14 anti-parallel β strands which form a β barrel (Court et al. 1995; Mannella et al. 1996). In contrast, all other Tom components are postulated to be anchored to the outer membrane by helical transmembrane segments. The import signals of these latter components were suggested to be located in the membrane anchor itself or in its flanking sequences (McBride et al. 1992; Cao and Douglas 1995; Rodriguez-Cousino et al. 1998).
The mitochondrial targeting signals in multi-topic membrane proteins, such as Tom40, are unknown. The information available regarding Tom40 import is that both receptor proteins, Tom20 and Tom70, appear to be involved in targeting, as antibodies against these two proteins inhibited Tom40 insertion (Keil et al. 1993). Despite its central role in the biogenesis of mitochondria, only little is known about the biogenesis of Tom40. Furthermore, as Tom40 is the main component of the TOM complex, any understanding of the assembly of this complex requires information on the insertion mechanism of this protein.
In the present study we investigated the mechanism by which Tom40 is targeted to the mitochondria, inserted into the outer membrane, and assembled into the TOM complex. Our results demonstrate that in a first stage Tom40 interacts with the surface receptor Tom20. Then it is integrated into preexisting TOM complexes. An NH2-terminally deleted form of Tom40 was unable to assemble into the TOM complex, whereas a COOH-terminally truncated form was assembled although with reduced stability. These results suggest the presence of a targeting signal in the internal part of Tom40 precursor and a signal required for assembly at the NH2 terminus. Denaturation of Tom40 before its in vitro import decreased the efficiency of insertion, indicating that Tom40 is probably inserted in a folded or partially folded conformation. Our results suggest a unique mechanism of targeting and membrane insertion of Tom40 that involves multiple steps of interaction with the translocation machinery.
Materials and Methods
Import of Preproteins into Isolated Mitochondria
Isolation of mitochondria from N. crassa was performed as described (Mayer et al. 1993). Radiolabeled preproteins were synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine (Amersham) after in vitro transcription by SP6 polymerase from pGEM4 vector containing the gene of interest. For urea treatment, two volumes of saturated ammonium sulfate solution were added to one volume of lysate. After precipitation on ice and centrifugation, the pellet was resuspended in 8 M urea, 10 mM MOPS-KOH, pH 7.2. For efficient denaturation, the urea-treated lysate was incubated at 25°C for 30 min before import reactions. The final concentration of urea in import reactions was always <350 mM. Import reactions were performed by incubation of radiolabeled preproteins with 30–50 μg mitochondria in import buffer (0.5% [wt/vol] BSA, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 2 mM ATP, 10 mM MOPS-KOH, pH 7.2) at the indicated temperature. Proteinase K (PK)1 treatment of samples was performed by incubation with the protease for 15 min on ice, followed by addition of 1 mM PMSF for 5 min. Import was analyzed by SDS-PAGE and the gels were viewed by autoradiography or quantified by phosphorimaging system (Fuji BAS 1500). Immunodecoration was according to standard procedures and was visualized by the ECL method (Amersham). In some experiments chemical amounts of precursor were used. The fusion protein, pSu9(1-69)-DHFR with a hexahistidinyl tag at the COOH terminus [pSu9(1-69)-DHFR-his6] was purified by Ni-NTA affinity chromatography from extracts of the E. coli strain DH5α carrying the pQE60-pSu9(1-69)-DHFR-his6 overexpression vector.
Construction of Tom40 Mutants
pGEM4-Tom40ΔN DNA and pGEM4-Tom40ΔC DNA were constructed by PCR amplification of the relevant DNA from pGEM4-Tom40. For pGEM4-Tom40ΔN, the upstream primer 5′-AGA AGA AAA GAA TTC ACC ATG TTC TCT GGC CTC CGC-3′ and the downstream primer 5′-CTC TAA GCT TTT AAA AGG GGA TGT TGA GG-3′ were used. For pGEM4-Tom40ΔC, the upstream primer 5′-AGA AGA AAA GAA TTC ACC ATG GCT TCG TTT TCC ACC-3′ and the downstream primer 5′-AGA AGA AAA AAG CTT CTA AAT GGA GAC GGA CAT GCC-3′ were used. Both PCR products were digested with EcoRI and HindIII and subcloned into pGEM4.
Cross-linking and Immunoprecipitation
For cross-linking experiments, radiolabeled precursors were incubated with isolated mitochondria under various conditions. After the import reaction, mitochondria were isolated and resuspended in import buffer followed by addition of the cross-linking reagents (Pierce) for 40 min at 0°C. The concentrations of the cross-linkers were 440 μM for disuccinimidyl glutarate (DSG) and 380 μM for m-maleimidobenzoyl-N-hydroxysulfo-succinimide ester (S-MBS). Excess cross-linker was quenched by the addition of 80 mM glycine, pH 8.0, and incubation for 15 min at 0°C. Aliquots were removed before and after addition of the cross-linking reagents. For immunoprecipitation, samples were dissolved in lysis buffer (1% SDS, 0.5% Triton X-100, 150 mM NaCl, 10 mM Tris-HCl, pH 7.2). After incubation for 5 min at 25°C, the lysed material was diluted 40-fold with lysis buffer lacking SDS. After a clarifying spin (15 min at 20,000 g), the supernatant was incubated with antibodies that were coupled to protein A–Sepharose beads.
Blue Native Gel Electrophoresis (BNGE)
Mitochondria (50–100 μg) were lysed in 50 μl digitonin buffer (1% digitonin, 20 mM Tris-HCl, 0.1 mM EDTA, 50 mM NaCl, 10% glycerol, 1 mM PMSF, pH 7.4). After incubation on ice for 10 min and clarifying spin (20 min, 22,000 g), 5 μl of sample buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM Bis-Tris, 500 mM 6-aminocaproic acid, pH 7.0) was added, and the mixture was analyzed by 6–13% gradient blue native gel (Schägger et al. 1994).
Results
The TOM Machinery Mediates Binding and Insertion of Tom40
To analyze the pathway of membrane insertion of Tom40, radiolabeled precursor was synthesized in vitro and incubated with mitochondria isolated from N. crassa. The formation of two specific proteolytic fragments (26 and 12 kD) upon treatment of mitochondria with PK was used as a criterion for correct insertion of the imported protein (Künkele et al. 1998b). The intensity of the 26-kD fragment (corrected for the reduction in the number of radiolabeled methionines) served to quantify the amount of inserted Tom40.
Efficient binding and insertion occurred at 25°C (Fig. 1 A). At 0°C the level of binding was moderately lower; however, only a minor fraction of the precursor was inserted into the membrane (Fig. 1 A). Pretreatment of mitochondria with trypsin to remove the exposed parts of the surface receptors resulted in an ∼50% reduction of binding and an ∼80–90% reduction of insertion of Tom40 (Fig. 1 A). The addition of apyrase, which hydrolyzes ATP, reduced the level of bound and of inserted Tom40 precursor to a similar extent. When Tom40 was imported in two steps, first binding at 0°C and then chase at 25°C, external ATP was not required in the second step (see below, Fig. 5). Thus, ATP appears to be required for release of the precursor from cytosolic factors rather than for the insertion step.
Figure 1.
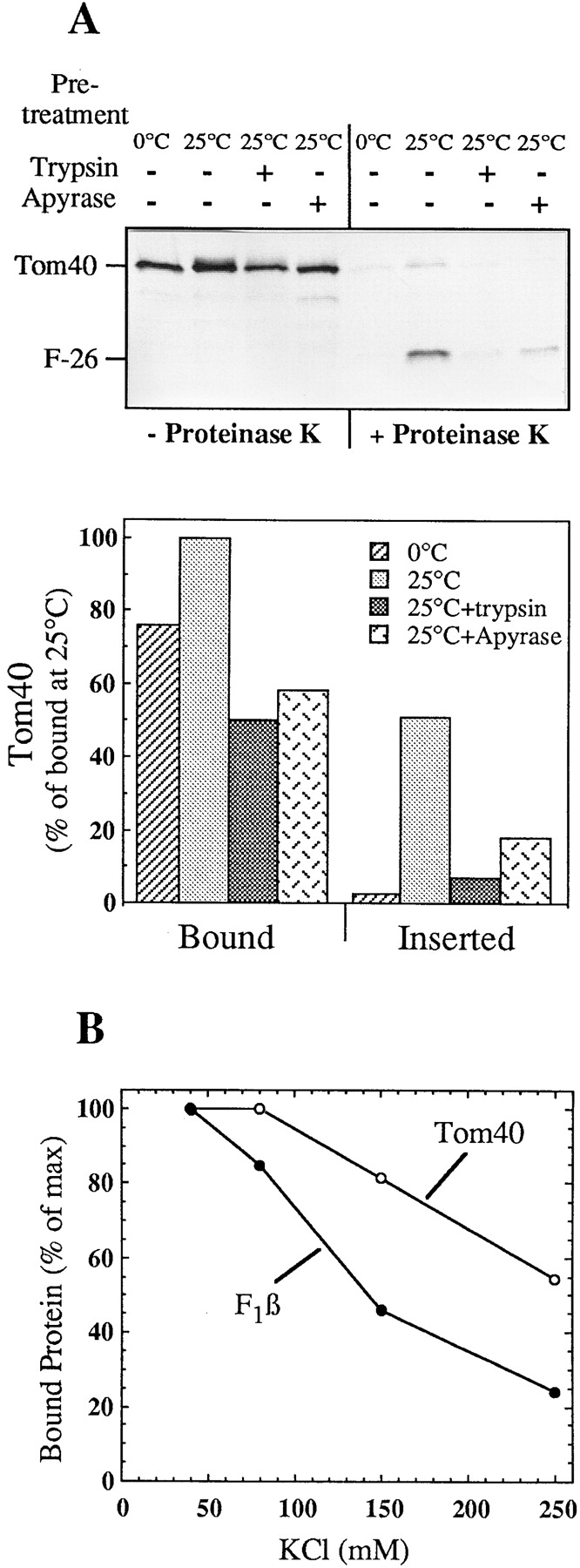
Insertion of Tom40 into the outer membrane of isolated mitochondria. (A) Import of the precursor of Tom40. Radiolabeled precursor was synthesized in reticulocyte lysate in the presence of [35S]methionine and incubated with isolated mitochondria for 15 min at 0°C or 25°C. When indicated, mitochondria were pretreated for 15 min at 0°C with either trypsin (50 μg/ml) or apyrase (0.5 U). After import each sample was divided into halves and one of them was treated with 160 μg/ml PK for 15 min at 0°C (+Proteinase K). Samples were analyzed by SDS-PAGE and autoradiography. F-26 is the characteristic NH2-terminal fragment of the endogenous Tom40 formed after PK treatment. In the lower panel the results are quantified and presented as a percentage of bound material at 25°C. Bound indicates total precursor without PK treatment. Inserted indicates a 26-kD band corrected for the loss of methionine residues. (B) Influence of salt on the import of Tom40. Radiolabeled precursors of Tom40 and F1β were imported into isolated mitochondria for 40 min at 0°C in import buffer containing the indicated concentrations of KCl. The total bound protein was quantified and plotted against the salt concentration.
Figure 5.


Intermediates on the insertion pathway of Tom40. (A) A trypsin-protected intermediate is formed at low temperature. Tom40 precursor was incubated with isolated mitochondria at 25°C or 0°C for the indicated periods of time. Then trypsin (60 μg/ml) or PK (200 μg/ml) was added for 15 min at 0°C. The trypsin protected material and formation of the 26-kD fragment resulting from PK treatment were analyzed. (B) The low temperature intermediate can be chased into the fully assembled protein. Radiolabeled precursor was incubated for 5 min at 0°C with isolated mitochondria. These were either treated with trypsin (120 μg/ml for 7 min) or centrifuged and resuspended in import buffer. The indicated samples were treated with PK immediately or after further incubation for 10 min at 0°C or 25°C. Protein bands corresponding to the 26-kD fragment were quantified and presented (after correction for loss of methionine residues) as a percentage of the bound protein in lane 1. (C) Tom40 binds initially as a monomer followed by the formation of a high molecular weight intermediate, I. Radiolabeled precursor was incubated at 0°C or 25°C with 50 μg mitochondria for the indicated periods of time. The indicated samples were treated with trypsin (80 μg/ml) and all samples were reisolated and solubilized in a buffer containing 1% digitonin. After clarifying spin, the solubilized mitochondria were loaded on a blue native gel. For detection of the endogenous TOM proteins, antibodies against Tom6, Tom22, and Tom40 were used. The three main stages of import are indicated: M, bound monomer; I, high molecular weight intermediate; and A, assembled TOM core complex. Unproductive material is indicated by an asterisk (upper panel). The bands corresponding to stages M, I, and A were quantified and are presented as a function of time of incubation (lower panels). (D) Intermediate I accumulated at 0°C can be chased to the assembled form by incubation at a higher temperature. Tom40 precursor was incubated with mitochondria for 10 min on ice followed by centrifugation and resuspension of mitochondria in import buffer (2 steps). The sample was divided into three aliquots that were incubated further for 20 min at either 0°C, 10°C, or 25°C. At the end of the second incubation period the mitochondria were centrifuged and solubilized in a buffer containing 1% digitonin. For comparison, precursor was imported for 20 min at 0°C and 25°C (1 step).
The binding efficiencies of Tom40 (and of F1β, a matrix-destined preprotein, for comparison) depended on the salt concentration. Binding of Tom40 decreased with increasing salt concentrations, although to a lesser extent than that of F1β, which bears a typical mitochondrial presequence (Fig. 1 B). The lower salt-sensitivity of the binding of Tom40 may suggest that in its binding, other forces like hydrophobic interactions are also involved.
To identify TOM components that interact with Tom40 precursor during binding and insertion, we performed chemical cross-linking experiments. Radiolabeled Tom40 was incubated with mitochondria under conditions that resulted in the formation of either a low temperature intermediate or the fully inserted protein. The reagents DSG or S-MBS were then added, and the cross-linking adducts were characterized and compared with cross-linking adducts of the endogenous Tom40 under the same conditions. Using DSG, a Tom40 dimer that contains the newly imported precursor was formed at 0°C, and with higher efficiency at 25°C (Fig. 2 A). This corresponds to an enhanced rate of assembly at 25°C (see Fig. 1 and Fig. 5). The dimer is similar to the dimer formed by the endogenous Tom40 (Fig. 2 A and Rapaport et al. 1998b). Since the imported material was present at an extremely low molar ratio relative to the preexisting Tom40 we assume the dimer-sized band results from cross-linking of an imported Tom40 to an endogenous Tom40. Thus, already at 0°C there appears to be an interaction between Tom40 precursor and endogenous Tom40 of the TOM complex. When S-MBS was added to the 0°C intermediate of Tom40, a unique cross-linking adduct was formed. This product was identified by immunoprecipitation as an adduct between Tom40 and Tom20 (Fig. 2 B). An adduct of Tom40 precursor with Tom20 was not observed upon import at 25°C nor an adduct with endogenous Tom40. These experiments indicate that Tom40 precursor interacts with Tom20 in initial stages of import but not after insertion.
Figure 2.

Tom40 interacts with Tom20 on its insertion pathway. (A) Radiolabeled Tom40 precursor was incubated with isolated mitochondria for 15 min at 0°C or 25°C. The mitochondria were reisolated, and divided into three aliquots. One aliquot was left on ice (−) while to the other two, the chemical cross-linkers DSG or S-MBS, were added for 40 min on ice. The cross-linking reagents were quenched with 80 mM glycine, pH 8.0, and the mitochondria were pelleted and loaded on SDS-PAGE. The gel was analyzed by autoradiography for detection of imported protein or immunostained with antibodies against Tom40 to detect the endogenous Tom40. (B) Identification of the Tom40-Tom20 adduct by immunoprecipitation. Radiolabeled Tom40 was incubated with isolated mitochondria for 15 min at 0°C. After reisolation, the cross-linker S-MBS was added (see above), and mitochondria were pelleted and solubilized with SDS- and Triton X-100–containing buffer. Aliquots were subjected to immunoprecipitation with antibodies against Tom20 or antibodies derived from preimmune serum. The immunoprecipitates were solubilized in sample buffer and analyzed as above.
Does Tom40 use the general insertion pore of the TOM complex for insertion? The protein conducting pore was blocked by accumulating chemical amounts of a translocation intermediate of the fusion protein pSu9(1-69)-DHFR in the presence of methotrexate, which stabilizes DHFR in a folded conformation (Ungermann et al. 1994; Dekker et al. 1997). This import intermediate, under the conditions of the experiment, spanned both mitochondrial membranes. Import of Tom40 into mitochondria containing this arrested intermediate was compared with import into control mitochondria (Fig. 3 A). Translocation of pF1β, which uses also the TIM machinery, was analyzed for comparison. Insertion of Tom40 was reduced by 75% and import of F1β by ∼95% as compared with the control (Fig. 3 A). Import of F1β was inhibited to a higher extent because the amounts of pSu9(1-69)-DHFR used were sufficiently high to saturate the Tim23-17 channels; they could not, however, block all the TOM channels which are present in higher amounts than TIM channels (Dekker et al. 1997; Sirrenberg et al. 1997). Next, we wanted to exclude the possibility that the insertion of Tom40 is blocked by the chemical amounts of the precursor at the level of the import receptors. To that end, mitochondria were first treated with trypsin to remove the exposed parts of the receptors, and then the effect of adding chemical amounts of pSu9-DHFR on insertion of Tom40 was determined. The proteolytic treatment resulted in reduced levels of Tom40 insertion (∼20% compared with control). However, also under these conditions of import bypassing the receptors pSu9-DHFR competed for insertion of Tom40 (Fig. 3 B). We conclude that Tom40 is using the general insertion pore for its import into the outer membrane.
Figure 3.
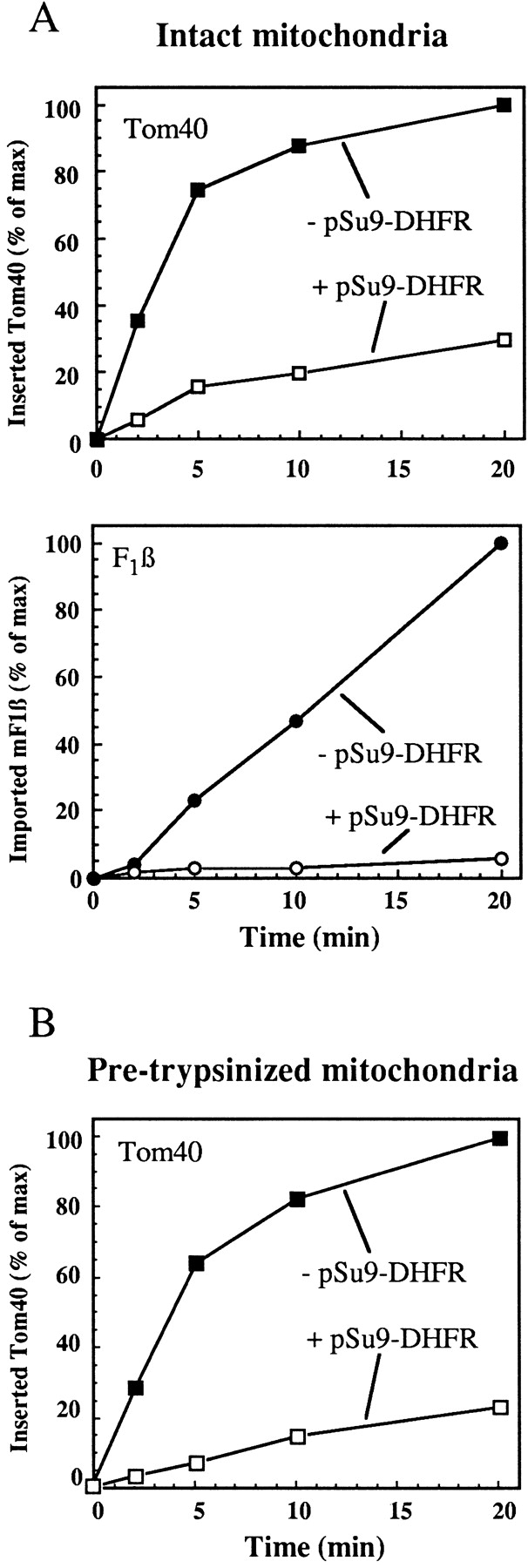
Insertion of Tom40 is inhibited by blocking the translocation pore with import intermediates. (A) Mitochondria in import buffer containing 1 μM methotrexate, 1 mM NADPH, and 2 mM NADH were incubated for 10 min at 0°C in the presence or absence of 3 μM of pSu9(1-69)-DHFR. Radiolabeled precursors of Tom40 or F1β were then added and further incubated at 15°C for the indicated periods. At the end of import, PK (100 μg/ml) was added and imported Tom40 (as 26-kD fragment) and mature F1β (mF1β) were quantified. (B) Mitochondria in import buffer containing 36 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP), 2 mM ATP, and 2 mM NADH were treated with trypsin (50 μg/ml) for 15 min at 0°C. The mitochondria were incubated for 10 min on ice in the presence or absence of 3 μM of pSu9(1-69)-DHFR. Radiolabeled precursor of Tom40 was then added and incubation continued at 25°C for the indicated time periods. At the end of import, PK (100 μg/ml) was added and imported Tom40 (as 26-kD fragment) was quantified.
Tom40 Is Inserted More Efficiently When It Is Partially Folded
An interesting aspect of the insertion of Tom40 is whether folding, at least partial folding, precedes or follows insertion into the outer membrane. We denatured Tom40 precursor by treating it with 8 M urea, and determined the efficiency of insertion in comparison to that of untreated precursor. The urea treatment strongly reduced the level of insertion (Fig. 4A and Fig. B). In control experiments, treatment of mitochondria with urea did not impair the insertion of native Tom40 precursor (not shown), eliminating the possibility that the reduced insertion in the case of urea-treated Tom40 was caused by destructive effects of urea on the mitochondrial import machinery. To exclude the possibility that this was the result of enhanced aggregation of the urea-treated precursor, we allowed the denatured material to refold in the presence of 33% reticulocyte lysate and only then added mitochondria. This treatment largely restored the ability of the precursor to become properly inserted (Fig. 4 A). As aggregation is usually a nonreversible process, these results suggest that most of the urea-treated precursor is not aggregated. In fact, upon BNGE, Tom40 precursor treated or untreated with urea migrated as a monomer (data not shown). Furthermore, the initial binding of Tom40 precursor to mitochondria is faster than aggregation of Tom40 upon dilution out of 8 M urea. Therefore, productive binding can occur before aggregation.
Figure 4.

Import of denatured Tom40 precursor. (A) Denaturation of Tom40 precursor reduces the insertion efficiency in reversible manner. Radiolabeled native Tom40 precursor or urea-treated precursor (3 μl each) was added to either 50 μg mitochondria in 100 μl import buffer containing 10% glycerol (3 μl reticulocyte lysate/100 μl reaction volume), or to a mixture of 60 μl import buffer (containing 10% glycerol) with 30 μl reticulocyte lysate for 20 min at 0°C, followed by the addition of 50 μg mitochondria (33 μl reticulocyte lysate/100 μl reaction volume). In both cases samples were incubated with mitochondria for 20 min at 15°C followed by reisolation by centrifugation, resuspension in import buffer, and dividing the samples into two aliquots. One was left on ice while the other was treated with PK. (B) Denaturation with urea has an opposite effect on import of Tom40 as compared with import of pSu9(1-45)-DHFR. Radiolabeled native or urea-denatured Tom40 and pSu9(1-45)-DHFR precursors were coincubated with 50 μg mitochondria at 15°C. After the indicated times, PK was added followed by centrifugation of the mitochondria and analysis of the proteins by SDS-PAGE (left panel). For comparison and for identification of the bands observed in the left panel, the native precursors were imported separately into mitochondria at 25°C and half of each reaction was treated with PK (right panel). The bands from the coimport experiment corresponding to the 26-kD fragment (F-26) of Tom40 and to the mSu9(1-45)-DHFR were quantified and presented as a percentage of the input (lower panel).
In the case of pSu9(1-45)-DHFR, a preprotein that is destined to the matrix and contains a presequence, denaturation with urea had a strong stimulating effect on import (Fig. 4 B). Hence, we conclude that for efficient insertion Tom40 must be folded, at least partially, when it interacts with the translocation machinery. This is in contrast to preproteins imported into the matrix where unfolding is a rate-limiting step (Eilers et al. 1988; Matouschek et al. 1997; Gaume et al. 1998; Rapaport et al. 1998a).
Tom40 Forms Translocation Intermediates
A further step in unraveling the insertion process of Tom40 was to characterize intermediates along the pathway and to address the question of whether the newly imported Tom40 becomes integrated into preexisting TOM complexes. The endogenous Tom40 in isolated mitochondria is resistant to added trypsin while PK cleaves it into two characteristic fragments (Kiebler et al. 1990; Künkele et al. 1998b). Upon import of Tom40 at 25°C the kinetics of acquisition of trypsin resistance was very similar to the kinetics of formation of the characteristic PK fragments (Fig. 5 A). At 0°C, however, there was no formation of the PK fragments while partial resistance to trypsin was acquired. This observation suggests the formation of an intermediate at 0°C, which is partially inserted into the outer membrane but not fully assembled. To demonstrate that this intermediate was a productive one we tried to chase it into the assembled species (Fig. 5 B). Tom40 precursor was incubated with mitochondria at 0°C for a short time and treatment with trypsin was performed. When the mitochondria were incubated in a second stage at 25°C, the 26-kD fragment could be generated by addition of PK. For comparison, the chase of surface-bound Tom40 precursor was also studied (Fig. 5 B). In this case, after incubation with the precursor at 0°C mitochondria were not treated with trypsin and therefore most of the precursor was present at the surface. This surface Tom40 intermediate was efficiently chased to inserted Tom40 (Fig. 5 B). Since trypsin-sensitive cytosolic domains of the TOM receptors support efficient insertion of Tom40 (see Fig. 1), the chase efficiency of trypsin-protected intermediate was lower than that of surface-bound precursor. Hence, the low temperature intermediate that is already partially inserted can be chased, although with low efficiency, into the fully inserted species.
The insertion pathway of Tom40 was analyzed further by BNGE. Radiolabeled Tom40 was incubated with mitochondria at 0°C or 25°C for various time periods. At the end of the import reactions the mitochondria were reisolated, solubilized in buffer containing 1% digitonin, and subjected to BNGE. Tom40 precursor bound initially at both 0°C and 25°C in a manner which resulted, under the conditions of the BNGE, in dissociation from the complex and migration as a monomer (abbreviated as M) (Fig. 5 C). A stable high molecular weight intermediate (abbreviated as I) was formed with slower kinetics at both temperatures. Whereas the monomer was completely trypsin-sensitive, the intermediate was partially trypsin-resistant. At 25°C, but not at 0°C, assembly into a complex (abbreviated as A) of the size of the authentic TOM complex was observed. Mitochondria were solubilized also with stronger detergents, such as Triton X-100 or dodecyl maltoside, instead of digitonin, to analyze the stability of the interaction of precursor with the TOM complex. Both detergents led to dissociation of the intermediate (I) but the assembled precursor was contained in the authentic core TOM complex (data not shown). These results suggest that the precursor in the intermediate state is interacting only loosely with the TOM machinery. The authentic TOM core complex was identified by immunodecoration with antibodies against TOM components, Tom6, Tom22, and Tom40 (Fig. 5 C). The vast majority of TOM complex, as analyzed by BNGE, did not contain the receptors Tom20 and Tom70 (data not shown). A similar loss of receptors from the yeast TOM core complex has been reported (Dekker et al. 1998). The monomer (M) and intermediate (I) species have the characteristics of true intermediates as demonstrated by kinetic analysis (Fig. 5 C). In contrast, the material indicated by an asterisk in Fig. 5 C appears to represent a nonproductive species, perhaps an artificial dimer. Based on the following observations we propose that the high molecular weight intermediate (I) contains, in addition to the precursor protein, endogenous TOM components: (a) small amounts of Tom40 and Tom6 were observed in a complex with a size identical to that of the high molecular weight intermediate (data not shown); (b) the band of the bound radioactive Tom40 monomer was not detected by immunodecoration, therefore the decorated Tom40 which was described above represents endogenous protein; and (c) a kinetic intermediate with identical molecular weight was observed upon import of Tom22 (data not shown).
To further establish band I as a true kinetic intermediate, a chase experiment was performed (Fig. 5 D). In a first step the radiolabeled precursor was incubated with mitochondria at 0°C. After reisolation, the mitochondria were incubated in a second step at different temperatures. When the second incubation was performed at 25°C efficient chase to the assembled complex was observed. In contrast, when the second step was performed again at 0°C only the high molecular weight intermediate (I) was observed. We suggest, therefore, that the main steps in the assembly of Tom40 are: (a) surface-bound monomer involving interaction with the receptor Tom20; (b) association of the precursor with the endogenous TOM complex followed by a precursor-induced conformational and/or structural change in the complex (resulting in higher mobility upon BNGE); and (c) assembly of Tom40 precursor into the TOM complex.
The NH2-terminal Segment Is Crucial for Assembly of Tom40
Tom40, like all outer membrane proteins, does not contain a cleavable targeting sequence. To find out whether the NH2- and the COOH-terminal portions contain information for targeting and insertion, we constructed two Tom40 variants. In Tom40ΔN residues 1–60 were deleted, while in the second, Tom40ΔC, the COOH-terminal residues 329–349 were removed. Both deleted segments were postulated to reside in the intermembrane space as soluble domains (Court et al. 1995). These variants were compared with the wild-type precursor with regard to their import into mitochondria. The amounts of total precursor associated with mitochondria were similar in all cases. Wild-type and both mutated forms were recovered in the membrane pellet after carbonate extraction (data not shown). Thus, both mutated forms were targeted to mitochondria, and neither the COOH- nor the NH2-terminal domain appeared to contain essential or exclusive targeting information.
To investigate the structural requirements for insertion and assembly of Tom40, the COOH- and NH2-terminally truncated variants were analyzed after import into isolated mitochondria. Tom40ΔC and, to a higher extent Tom40ΔN, were much more sensitive to trypsin added to intact mitochondria than wild-type Tom40 (Fig. 6 A). Hence, although the variant precursors are able to become inserted into the outer membrane, the deleted segments contain information required for acquisition of native-like (trypsin-resistant) conformation.
Figure 6.

Tom40 with deletions at the NH2-terminal (ΔN) or the COOH-terminal (ΔC) has decreased stability after insertion. (A) Reduced trypsin resistance of inserted variants. Radiolabeled Tom40 and mutated precursors were incubated with mitochondria at 25°C. After the indicated time periods trypsin (133 μg/ml) was added and mitochondria were isolated by centrifugation and subjected to SDS-PAGE. The bands corresponding to trypsin-protected precursors were quantified and are shown as percentage of input. (B) Segments of the variant precursors can insert correctly into the outer membrane. Radiolabeled Tom40 and mutated precursors were incubated with isolated mitochondria for 20 min at 25°C. Samples were treated with PK, mitochondria reisolated by centrifugation, and 10% were subjected to SDS-PAGE (Total). The rest was dissolved in buffer containing SDS and Triton X-100, split into three aliquots, and subjected to immunoprecipitation with antibodies against an NH2-terminal and a COOH-terminal epitope, or from preimmune serum (PIS). The two fragments, F26 and F12, resulting from PK treatment of Tom40 are indicated.
Is the Tom40 precursor inserted in a concerted manner or does this occur by a sequential pathway whereby domains insert independently of each other? After import of the precursor proteins and treatment with PK, immunoprecipitation was performed with antibodies against NH2- or COOH-terminal peptides of Tom40. In the case of the wild-type Tom40, the 26-kD fragment was recognized by the antibody against the NH2-terminal epitope, and the 12-kD fragment by the antibody raised against the COOH-terminal peptide (Fig. 6 B). Deletion of the COOH-terminal domain did not prevent the formation of the 26-kD fragment of the NH2-terminal part. Similarly, the formation of the typical 12-kD fragment of the COOH-terminal was observed even when the NH2-terminal was deleted (Fig. 6 B). If Tom40ΔN was inserted properly in the outer membrane one would expect the formation of a 19- instead of 26-kD band upon treatment with PK. Such a fragment was not observed, indicating impaired folding of this variant. These results suggest that some domains of Tom40 can be inserted despite an overall altered conformation of the entire molecule. Furthermore, none of the terminal segments contains exclusive information for the insertion of Tom40 precursor.
Assembly of the variant Tom40 precursors into the endogenous TOM complex was studied by coimmunoprecipitation and BNGE. To ensure, in the immunoprecipitation procedure, that imported molecules would not be recognized directly by the antibodies but only upon their interaction with endogenous Tom40 molecules, antibodies raised against the missing domain in the truncated proteins were used. To exclude the presence of steady-state levels of assembly intermediates of the TOM complex, we compared import into mitochondria from normally grown Neurospora with import into mitochondria from a Neurospora culture that had received cycloheximide (CHX) during the last 90 min of growth. CHX blocks the synthesis of new proteins, therefore treatment of cells before isolation of the mitochondria minimizes the possibility for the presence of assembly-intermediates in the mitochondrial outer membrane that might react with Tom40 imported in vitro. Variant precursors of Tom40 were imported into mitochondria from CHX-treated or -untreated cells and immunoprecipitation with antibodies against NH2 or COOH termini of Tom40 was performed (Fig. 7 A). Both NH2- and COOH-terminally truncated forms of Tom40 were observed to interact with endogenous molecules of Tom40. The amount of Tom40ΔN coprecipitated with preexisting TOM complexes was much lower than that of Tom40ΔC. Further analysis by BNGE suggested that the Tom40ΔN was not integrated into the fully assembled TOM complex but rather reached only the intermediate stage (I) (Fig. 7 B). In contrast, Tom40ΔC was assembled with similar efficiency as the full length precursor. Assembly of Tom40ΔN was further analyzed by coimmunoprecipitation with antibodies against Tom22 and Tom6. These subunits interact with Tom40 in the assembled TOM complex. Imported native Tom40 was efficiently coimmunoprecipitated, but only minor amounts of Tom40ΔN were precipitated by these two antibodies (Fig. 7 C). These results indicate that the NH2-terminal domain contains crucial information for the correct assembly of Tom40.
Figure 7.

The NH2-terminal domain of Tom40 is essential for correct assembly. (A) The Tom40ΔN and the Tom40ΔC precursors were incubated with isolated mitochondria from normal culture or from a culture to which CHX (150 μg/ml) was added 90 min before isolation of mitochondria (+CHX). After incubation for 20 min at 25°C the mitochondria were reisolated and one sixth of the material was loaded on the gel (Total). The rest was solubilized with 0.5% digitonin, halved, and subjected to immunoprecipitation with the indicated antibodies or with antibodies from preimmune serum (PIS). (B) Tom40ΔN cannot be fully assembled into the Tom complex. Radiolabeled precursors were incubated with intact mitochondria for 20 min at 25°C. Further treatment and analysis by BNGE was as described in Fig. 5 C. Fully assembled material and the high molecular intermediate are indicated by A and I, respectively. (C) Tom40ΔN is not stably associated with Tom22 and Tom6. Tom40 and Tom40ΔN precursors were incubated with mitochondria for 20 min at 25°C and mitochondria were reisolated. One aliquot was directly analyzed by SDS-PAGE, the rest was solubilized with 0.5% digitonin, split into three aliquots which were subjected to immunoprecipitation with antibodies against Tom22, or Tom6, or with antibodies from preimmune serum (PIS). PIS could not precipitate the precursors (data not shown).
Discussion
We have analyzed the pathway of insertion into the outer membrane of Tom40, the major component of the TOM machinery. A working model for this process is presented (Fig. 8). Efficient insertion of Tom40 requires ATP and a (partially) folded state. Cytosolic chaperones are probably involved in keeping Tom40 in a translocation-competent state. The hydrolysis of ATP may provide the energy required to release the chaperones from the precursor (Fig. 8). ATP requirement for import was reported for most outer membrane proteins (Shore et al. 1995).
Figure 8.

Working model of the insertion pathway of Tom40. Tom40 is presented to the mitochondria in a (partially) folded state and is initially recognized on the surface by the receptor Tom20. At this or a previous stage it is released in an ATP-dependent process from its interaction with cytosolic factors. The surface-bound Tom40 is only loosely attached to the TOM complex and after solubilization with mild detergent can be detected in monomeric state (M). In the next stage, initial insertion of the precursor into the translocation pore occurs (I). By this step the precursor acquires a state partially protected from externally added trypsin. The processes up to this point can take place rather efficiently also at 0°C. The next step, full assembly into the TOM complex (A), requires higher temperature and occurs when the NH2 terminus of Tom40 is intact.
In most translocation systems the substrate proteins are translocated in a largely unfolded state (Schatz and Dobberstein 1996). Mitochondrial preproteins that contain a tightly folded domain were shown to become stalled, spanning across the mitochondrial import machinery (Ungermann et al. 1994; Horst et al. 1995; Dekker et al. 1997). Accordingly, denaturation of precursor proteins was found to improve translocation efficiency (Vestweber and Schatz 1988; Matouschek et al. 1997; Rapaport et al. 1998a). The insertion mechanism of Tom40 appears to involve rather different folding requirements. Our results suggest that Tom40 has to be at least partially folded in order to become efficiently inserted into the outer membrane. Tom40 was postulated to be composed of a series of β sheets that form a β barrel. Insertion of β barrel proteins may involve a concerted partitioning of β strands into the membrane; thereby sufficient hydrophobic character would be available to favor bilayer integration (Singer 1990). Similarly, bacterial porins, established β barrel proteins, were proposed to be at least partially folded before they insert into the outer membrane (Eppens et al. 1997). Like other outer membrane proteins Tom40 is synthesized without an NH2-terminal presequence. Which part of the Tom40 molecule contains the targeting and sorting information? Our results propose the targeting information of Tom40 is not exclusively present at the NH2 and the COOH termini. In the case of proteins that are predicted to traverse the outer membrane only once, the targeting information was localized to a single contiguous sequence (McBride et al. 1992; Cao and Douglas 1995; Rodriguez-Cousino et al. 1998). In the case of Tom40, however, the requirement for a (partially) folded state suggests targeting information may be composed of discontinuous sites in a folded domain structure.
The basic organization of Tom40 appears to be a dimer (Dekker et al. 1998; Rapaport et al. 1998b). We propose that Tom40 is imported as a monomer and dimerization is taking place only after insertion into the outer membrane. This notion is supported by the following observations: (a) using the chemical cross-linker DSG we observed Tom40 dimer after import into mitochondria but not in solution (Rapaport et al. 1998b and unpublished results); (b) radiolabeled Tom40 migrated in a blue native gel system as a monomer which, upon insertion, was converted to a higher molecular weight species. Bacterial porins were also suggested to be inserted as a monomer followed by trimerization in the membrane (Van Gelder et al. 1994; Surrey et al. 1996).
Tom40 utilizes the TOM complex for its insertion, like other TOM components such as Tom22 and Tom70 (Schlossmann and Neupert 1995; Court et al. 1996). An early insertion intermediate of Tom40 is formed even at low temperature. This intermediate is loosely attached to the TOM complex and directly interacts with Tom20, a surface receptor of the TOM machinery. Thus, Tom40, like presequence-containing preproteins and other outer membrane proteins (Ramage et al. 1993; Harkness et al. 1994), uses Tom20 as the initial binding partner on the surface of the mitochondria (Fig. 8). The involvement of Tom20 in the insertion of Tom40 is supported by previous studies; antibodies against Tom20 were found to inhibit the import of Tom40 (Keil et al. 1993), and the efficiency of import of Tom40 into mitochondria from a Tom20-deficient strain was highly reduced (Harkness et al. 1994).
A next step in the assembly process of Tom40 is association in a rather stable manner with a high molecular weight complex. The structure formed in this way contains components of the endogenous TOM machinery like Tom40 and Tom6. In this intermediate a Tom40 precursor is partially inserted into the membrane but not yet assembled into the preexisting TOM complex (Fig. 8). This is in agreement with observations on the insertion pathway of a Tom40 variant with a truncated NH2 terminus. This variant can form the intermediate, but cannot go further; still it can become inserted into the outer membrane. Interestingly, porin mutants with a marked instability of the trimeric state were still able to become inserted into the bacterial outer membrane (Fourel et al. 1994). Such a lack of correlation between localization and stabilization may indicate that the signals for these processes do not necessarily overlap.
The newly inserted Tom40 is finally assembled into preexisting TOM complexes (Fig. 8). This last step is blocked at low temperatures and requires the NH2-terminal segment of Tom40. It seems likely that the TOM complex initially releases a newly imported Tom40 into the outer membrane which then assembles with other Tom40 molecules into the TOM complex. A minor population of partial complexes which is in equilibrium with fully assembled TOM complexes could serve as sites of integration of new Tom40 species. On the other hand, a direct insertion of the Tom40 intermediate into the TOM complex cannot be excluded at present. Taken together, our results suggest that Tom40 follows a unique pathway in its insertion pathway into the outer membrane.
Acknowledgments
The excellent technical assistance of Petra Heckmeyer is gratefully acknowledged. We thank Dr. R. Lill for helpful discussions.
This research was supported by the Sonderforschungsbereich 413 of the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and a postdoctoral fellowship of the E.U.-Training and Mobility of Researchers (to D. Rapaport).
Footnotes
1.used in this paper: BNGE, blue native gel electrophoresis; CHX, cycloheximide; DSG, disuccinimidyl glutarate; PK, proteinase K; S-MBS, m-maleimidobenzoyl-N-hydroxysulfo-succinimide ester
References
- Cao W., Douglas M.G. Biogenesis of ISP6, a small carboxyl-terminal anchored protein of the receptor complex of the mitochondrial outer membrane. J. Biol. Chem. 1995;270:5674–5679. doi: 10.1074/jbc.270.10.5674. [DOI] [PubMed] [Google Scholar]
- Court D.A., Lill R., Neupert W. The protein import apparatus of the mitochondrial outer membrane. Can. J. Bot. 1995;73:193–197. [Google Scholar]
- Court D.A., Nargang F.E., Steiner H., Hodges R.S., Neupert W., Lill R. Role of the intermembrane space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol. Cell. Biol. 1996;16:4035–4042. doi: 10.1128/mcb.16.8.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker P.J.T., Martin F., Maarse A.C., Bömer U., Müller H., Guiard B., Meijer M., Rassow J., Pfanner N. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO (Eur. Mol. Biol. Org.) J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker P.J.T., Ryan M.T., Brix J., Müller H., Hönlinger A., Pfanner N. Preprotein translocase of the outer mitochondrial membranemolecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 1998;18:6515–6524. doi: 10.1128/mcb.18.11.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Hwang S., Schatz G. Unfolding and refolding of a purified precursor protein during import into isolated mitochondria. EMBO (Eur. Mol. Biol. Org.) J. 1988;7:1139–1145. doi: 10.1002/j.1460-2075.1988.tb02923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppens E.F., Nouwen N., Tommassen J. Folding of a bacterial outer membrane protein during passage through the periplasm. EMBO (Eur. Mol. Biol. Org.) J. 1997;16:4295–4301. doi: 10.1093/emboj/16.14.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel D., Bernadac A., Pagès J.-M. Involvement of exposed polypeptide loops in trimeric stability and membrane insertion of Escherichia coli OmpF porin. Eur. J. Biochem. 1994;222:625–630. doi: 10.1111/j.1432-1033.1994.tb18905.x. [DOI] [PubMed] [Google Scholar]
- Gaume B., Klaus C., Ungermann C., Guiard B., Neupert W., Brunner M. Unfolding of preproteins upon translocation across membranes of mitochondria. EMBO (Eur. Mol. Biol. Org.) J. 1998;22:6497–6507. doi: 10.1093/emboj/17.22.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B., Schatz G. Import of proteins into mitochondria. Annu. Rev. Genet. 1991;25:21–44. doi: 10.1146/annurev.ge.25.120191.000321. [DOI] [PubMed] [Google Scholar]
- Harkness T.A., Nargang F.E., van der Klei I., Neupert W., Lill R. A crucial role of the mitochondrial protein import receptor MOM19 for the biogenesis of mitochondria. J. Cell Biol. 1994;124:637–648. doi: 10.1083/jcb.124.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haucke V., Schatz G. Import of proteins into mitochondria and chloroplasts. Trends Cell Biol. 1997;7:103–106. doi: 10.1016/S0962-8924(96)10052-0. [DOI] [PubMed] [Google Scholar]
- Hill K., Model K., Ryan M.T., Dietmeier K., Martin F., Wagner R., Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Horst M., Hilfiker R.S., Oppliger W., Schatz G. Dynamic interaction of the protein translocation systems in the inner and outer membranes of yeast mitochondria. EMBO (Eur. Mol. Biol. Org.) J. 1995;14:2293–2297. doi: 10.1002/j.1460-2075.1995.tb07223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil P., Weinzierl A., Kiebler M., Dietmeier K., Sollner T., Pfanner N. Biogenesis of the mitochondrial receptor complex. Two receptors are required for binding of MOM38 to the outer membrane surface. J. Biol. Chem. 1993;268:19177–19180. [PubMed] [Google Scholar]
- Kiebler M., Pfaller R., Söllner T., Griffiths G., Horstmann H., Pfanner N., Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature. 1990;348:610–616. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- Künkele K.-P., Heins S., Dembowski M., Nargang F.E., Benz R., Thieffry M., Walz J., Lill R., Nussberger S., Neupert W. The preprotein translocation channel of the outer membrane of mitochondria Cell. 93 1998. 1009 1019a [DOI] [PubMed] [Google Scholar]
- Künkele K.-P., Juin P., Pompa C., Nargang F.E., Henry J.-P., Neupert W., Lill R., Thieffry M. The isolated complex of the translocase of the outer membrane of mitochondria J. Biol. Chem. 273 1998. 31032 31039b [DOI] [PubMed] [Google Scholar]
- Lill R., Neupert W. Mechanisms of protein import across the mitochondrial outer membrane. Trends Cell Biol. 1996;6:56–61. doi: 10.1016/0962-8924(96)81015-4. [DOI] [PubMed] [Google Scholar]
- Mannella C., Neuwald A., Lawrence C. Detection of likely beta-strand regions in sequences of mitochondrial pore proteins using the Gibbs sampler. J. Bioenerg. Biomembr. 1996;28:163–169. doi: 10.1007/BF02110647. [DOI] [PubMed] [Google Scholar]
- Matouschek A., Azem A., Ratliff K., Glick B.S., Schmid K., Schatz G. Active unfolding of precursor proteins during mitochondrial protein import. EMBO (Eur. Mol. Biol. Org.) J. 1997;16:6727–6736. doi: 10.1093/emboj/16.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Lill R., Neupert W. Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J. Cell Biol. 1993;121:1233–1243. doi: 10.1083/jcb.121.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H.M., Millar D.G., Li J.M., Shore G.C. A signal-anchor sequence selective for the mitochondrial outer membrane. J. Cell Biol. 1992;119:1451–1457. doi: 10.1083/jcb.119.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Rassow J., van der Klei I., Neupert W. A dynamic model of the mitochondrial protein import machinery. Cell. 1992;68:999–1002. doi: 10.1016/0092-8674(92)90069-o. [DOI] [PubMed] [Google Scholar]
- Ramage L., Junne T., Hahne K., Lithgow T., Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO (Eur. Mol. Biol. Org.) J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Neupert W., Lill R. Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J. Biol. Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Mayer A., Neupert W., Lill R. Cis and trans sites of the TOM complex in unfolding and initial translocation of preproteins J. Biol. Chem. 273 1998. 8806 8813a [DOI] [PubMed] [Google Scholar]
- Rapaport D., Künkele K.-P., Dembowski M., Ahting U., Nargang F.E., Neupert W., Lill R. Dynamics of the TOM complex of mitochondria during binding and translocation of preproteins Mol. Cell. Biol. 18 1998. 5256 5262b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cousino N., Nargang F.E., Baardman R., Neupert W., Lill R., Court D.A. An import signal in the cytosolic domain of the Neurospora mitochondrial outer membrane protein Tom22. J. Biol. Chem. 1998;272:11527–11532. doi: 10.1074/jbc.273.19.11527. [DOI] [PubMed] [Google Scholar]
- Schägger H., Cramer W.A., von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schatz G., Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schlossmann J., Neupert W. Assembly of the preprotein receptor MOM72/MAS70 into the protein import complex of the outer membrane of mitochondria. J. Biol. Chem. 1995;270:27116–27121. doi: 10.1074/jbc.270.45.27116. [DOI] [PubMed] [Google Scholar]
- Shore G.C., McBride H.M., Millar D.G., Steenaart N.A.E., Nguyen M. Import and insertion of proteins into the mitochondrial outer membrane. Eur. J. Biochem. 1995;227:9–18. doi: 10.1111/j.1432-1033.1995.tb20354.x. [DOI] [PubMed] [Google Scholar]
- Singer S.J. The structure and insertion of integral membrane proteins in membranes. Annu. Rev. Cell Biol. 1990;6:247–296. doi: 10.1146/annurev.cb.06.110190.001335. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C., Endres M., Becker K., Bauer M.F., Walther E., Neupert W., Brunner M. Functional cooperation and stoichiometry of protein translocases of the outer and inner membranes of mitochondria. J. Biol. Chem. 1997;272:29963–29966. doi: 10.1074/jbc.272.47.29963. [DOI] [PubMed] [Google Scholar]
- Stuart R.A., Neupert W. Topogenesis of inner membrane proteins of mitochondria. Trends Biochem. Sci. 1996;21:261–267. [PubMed] [Google Scholar]
- Surrey T., Schmid A., Jähnig F. Folding and membrane insertion of the trimeric β-barrel protein OmpF. Biochemistry. 1996;35:2283–2288. doi: 10.1021/bi951216u. [DOI] [PubMed] [Google Scholar]
- Ungermann C., Neupert W., Cyr D.M. The role of Hsp70 in conferring unidirectionality on protein translocation into mitochondria. Science. 1994;266:1250–1253. doi: 10.1126/science.7973708. [DOI] [PubMed] [Google Scholar]
- Van Gelder P., De Cock H., Tommassen J. Detergent-induced folding of the outer-membrane protein PhoE, a pore protein induced by phosphate limitation. Eur. J. Biochem. 1994;226:783–787. doi: 10.1111/j.1432-1033.1994.00783.x. [DOI] [PubMed] [Google Scholar]
- Vestweber D., Schatz G. Point mutations destabilizing a precursor protein enhance its post-translational import into mitochondria. EMBO (Eur. Mol. Biol. Org.) J. 1988;7:1147–1151. doi: 10.1002/j.1460-2075.1988.tb02924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D., Brunner J., Baker A., Schatz G. A 42K outer-membrane protein is a component of the yeast mitochondrial protein import site. Nature. 1989;341:205–209. doi: 10.1038/341205a0. [DOI] [PubMed] [Google Scholar]



