Abstract
Advances in proteomic techniques have allowed the large-scale identification of phosphorylation sites in complex protein samples, but new biological insight requires an understanding of their in vivo dynamics. Here, we demonstrate the use of a stable isotope-based quantitative approach for pathway discovery and structure–function studies in Arabidopsis cells treated with the bacterial elicitor flagellin. The quantitative comparison identifies individual sites on plasma membrane (PM) proteins that undergo rapid phosphorylation or dephosphorylation. The data reveal both divergent dynamics of different sites within one protein and coordinated regulation of homologous sites in related proteins, as found for the PM H+-ATPases AHA1, 2 and 3. Strongly elicitor-responsive phosphorylation sites may reflect direct regulation of protein activity. We confirm this prediction for RbohD, an NADPH oxidase that mediates the rapid production of reactive oxygen species (ROS) in response to elicitors and pathogens. Plant NADPH oxidases are structurally distinct from their mammalian homologues, and regulation of the plant enzymes is poorly understood. On RbohD, we found both unchanging and strongly induced phosphorylation sites. By complementing an RbohD mutant plant with non-phosphorylatable forms of RbohD, we show that only those sites that undergo differential regulation are required for activation of the protein. These experiments demonstrate the potential for use of quantitative phosphoproteomics to determine regulatory mechanisms at the molecular level and provide new insights into innate immune responses.
Keywords: quantitative phosphoproteomics, protein phosphorylation, signal transduction, innate immunity, plasma membrane, mass spectrometry
Introduction
A portion of a plant’s resistance to potential bacterial pathogens is mediated by recognition of characteristic ‘non-self’ structures associated with invading microbes, so-called elicitors or pathogen-associated molecular patterns (PAMPs) (Zipfel and Felix, 2005). One such perception systems involves the recognition of a conserved portion of flagellin, the principal building block of bacterial flagella, by the FLS2 receptor kinase (Felix et al., 1999; Gómez-Gómez and Boller, 2000). Plants lacking FLS2 are more susceptible to virulent bacteria and are impaired in establishing induced resistance after pre-treatment with bacterial extracts or a flagellin-derived peptide (Zipfel et al., 2004). However, the pathway(s) leading from PAMP recognition to resistance remain poorly understood.
Protein phosphorylation is required for numerous PAMP responses, including production of ROS and regulation of ion fluxes (Chandra and Low, 1995; Felix et al., 1991). A MAP kinase pathway has been shown to be rapidly regulated in response to PAMP treatment (Asai et al., 2002), but links between kinase activation and the diverse cellular defence responses are still lacking (Peck, 2003). In particular, very few differentially phosphorylated proteins have been identified.
We previously described a directed proteomic approach based on in vivo pulse labelling with 33P and 2D PAGE to identify proteins that are rapidly phosphorylated in response to flagellin (Peck et al., 2001). Because the earliest, PM-localized elicitor responses are dependent on phosphorylation (Chandra and Low, 1995; Felix et al., 1991), we specifically targeted PM proteins in a parallel approach. Two differentially phosphorylated syntaxins, AtSYP122 (Nühse et al., 2003a) and AtSYP132 (M. Kalde and S.C.P., The Sainsbury Laboratory, unpublished data), were identified. Larger, more hydrophobic membrane proteins, however, were not accessible using the 2D PAGE technology. We therefore developed a peptide-based approach using immobilized metal ion affinity chromatography (IMAC) to enrich for phosphopeptides from complex protein mixtures (Nühse et al., 2003b). This strategy yielded directly the sequences of hundreds of in vivo phosphorylation sites of Arabidopsis PM proteins. These data, however, were qualitative and static. In order to study the PAMP-regulated changes in protein phosphorylation, a quantitative method was needed to compare different samples.
Here, we describe the isotopic labelling of peptides to achieve quantification of dynamic protein phosphorylation in Arabidopsis. The nature of the labelling technology makes it universally applicable both to different biological samples [cell cultures, differentiated tissues, subcellular fractions or (partially) purified samples] and different treatments or developmental stages. Applying it to the response of cultured cells to elicitation with flg22, we identified several new differentially phosphorylated proteins with a potential role in defence response, as well as differential phosphorylation of proteins previously associated with plant–pathogen interactions on the basis of genetic screens. We also confirmed that this strategy provides information about mechanisms of protein regulation. For the NADPH oxidase involved in the rapid production of ROS, RbohD, we confirmed that two phosphorylation sites that are strongly induced by elicitor treatment are required for its activation, while a site that was unchanged after elicitation could be mutated without effect on protein activity.
Results and discussion
Establishing criteria for quantitative comparisons
For quantitative phosphoproteomic experiments, stable isotope labelling with amino acids in cell culture (SILAC) has become the predominant method (Ong et al., 2002). In plant cells, however, labelling is relatively inefficient (Gruhler et al., 2005). Complete metabolic labelling with 15N via inorganic nitrate and/or ammonia is an alternative that is better suited to plants (Benschop et al., in press). However, 15N labelling and SILAC, like most alternative methods, are limited to comparisons of two or three samples at the most. A priori, this limitation makes the acquisition of data from time courses or simultaneous comparisons of responses in different genotypes more complicated and potentially less reproducible. As an alternative to these methods, we explored the use of isobaric tags for relative quantification (iTRAQ; Ross et al., 2004). This post-digestion labelling tags all tryptic peptides, which is crucial for the analysis of post-translational modifications. In addition, the four available tags allow quadruplex experiments such as a time course or internal repeats of biological treatments. Both of these strategies were utilized in the present studies (Figure 1).
Figure 1.
Schematic work flow for quantitative phosphoproteomics. Steps performed in parallel with control or treated samples are indicated with four individual block arrows in white or grey/black. All procedures after the pooling (indicated with asterisk) of iTRAQ labelled peptides are indicated with a single broad multicoloured arrow. Phosphopeptides were isolated either from labelled and pooled samples (a) or from larger amounts of unlabelled samples, followed by iTRAQ labelling (b). Note that ion-exchange fractionation is required for sample deconvolution, desalting before IMAC and post-iTRAQ labelling sample clean-up, which means that two rounds of ion-exchange fractionation are necessary in (b).
The quadruplex iTRAQ label was used either to analyse a time course comprising one control and three elicited samples (0, 3, 7 and 15 min after elicitation, experiments 1–3), or a pair of two independent experiments with one control and one elicited sample each (0 and 7 min, experiments 4/5 and 6/7; see Experimental procedures and Appendix S1). Labelling of peptides with the iTRAQ reagent was performed either before (Figure 1a; experiments 1–3) or after isolation of phosphopeptides from the digest (Figure 1b; experiments 4–7). Isolating phosphopeptides before iTRAQ labelling allowed a much larger input of material because one set of iTRAQ reagents labels a maximum of 4 × 100 μg protein digest. Using phosphopeptides isolated from as much as 2 mg protein digest per sample for each individual iTRAQ labelling reaction, we achieved more robust signals in the mass spectrometric analysis.
The phosphopeptide samples were very complex despite ion-exchange fractionation, and the limited overlap of the peptides identified in each experiment reflects undersampling during the LC–MS/MS analysis. Many of the peptides identified in the present study had not been found in our previous large-scale study (Nühse et al., 2003b), possibly because of different biases in the pre-fractionation technique (cation versus anion exchange).
Although parallel PM isolations for control and treated samples required multiple processing steps over several hours, we found that, in comparable experiments without phosphopeptide isolation, the distribution of treated versus control ratios was relatively narrow (Supplementary Figure S1), and larger than twofold changes in abundance of a peptide were significant at the 99% level. Phosphopeptides were distributed slightly more broadly (Supplementary Figure S1), which may reflect weak regulation of a large number of phosphorylation sites by the elicitor. We expected that additional parallel sample processing before pooling the iTRAQ-labelled peptides (Figure 1b) might further increase the variation of treated/control ratios. However, the increase in distribution of ratios only slightly affected the statistical confidence (Supplementary Figure S1, experiments 4–7). Therefore, post-IMAC labelling is a viable approach where the limits of LC–MS/MS sensitivity affect the coverage of the phosphoproteome.
To identify significantly induced phosphopeptides while accounting for a broad biological effect on the treated/control ratio distribution, we set the thresholds at P= 0.05 or P= 0.1, which correlates to an approximately twofold change in most experiments.
To define a phosphorylation site as either increasing or decreasing during the response, we required that the peptide be found to change significantly in at least two experiments and not show a contradicting accumulation pattern in any other experiment. For strongly induced sites, the ‘fold induction’ values often varied considerably, probably due to the rapid kinetics of the elicitor response (Felix et al., 1999; Nühse et al., 2000) and the challenge of rapidly harvesting large volumes of cell cultures. Some significantly induced peptides were found in only a single experiment or were consistently increased in elicited samples but below the conservative threshold (such as S1053 of callose synthase PMR4/GSL5, At4g03550; full data in Supplementary Table S1). Although these candidates appear promising, they require further experiments for validation.
Both previously known defence-related proteins and novel proteins contain strongly elicitor-induced phosphorylation sites
Based on the criteria described above, we identified 11 PM proteins undergoing differential phosphorylation (Table 1 and Figure 2). Some phosphopeptides with Mascot scores below the P= 0.05 significance threshold have been included where careful examination of the spectrum supported the database match (see Appendix S1). Spectra for all peptides in Table 1 are available in Supplementary Figure S2. With the exception of two H+-ATPase sites (see Figure 2 and Supplementary Table S2), no phosphorylation sites were consistently and significantly reduced in response to the elicitor. Among the proteins with induced phosphorylation sites (Table 1) were novel proteins and two with a recognized role in pathogen defence, the ABC transporter PDR8/PEN3 and the NADPH oxidase RbohD (see biological characterization of the differentially phosphorylated residues of RbohD, below). Although one might expect the FLS2 receptor kinase involved in transmitting the flg22 signal to be phosphorylated during elicitation, we did not observe any phosphopeptides from this protein. However, in targeted experiments analysing immunoprecipitated FLS2, we found a single phosphopeptide from the protein that could not be enriched by IMAC (data not shown). These experiments also raise caution that other regulated phosphopeptides might similarly have escaped detection as a result of different chromatographic properties.
Table 1.
Significantly elicitor-induced phosphorylation sites
| Gene identifier | Protein name | Identified peptide and phosphorylation site | Fold induction (7 min elicited versus control) |
|---|---|---|---|
| At1g59610 | Dynamin-related protein 2B (ADL3) | 829AAAASSWSDNSGTESSPR (1P) | 1.5–24* |
| At1g59870 | PDR8/PEN3 | 31NIEDIFSSGSRR (1P) | 6–8 |
| ABC transporter | 31NIEDIFSSGSR | 3–16 | |
| 43TQSVNDDEEALK | 4–14 | ||
| 823SLSTADGNR | 1.7–2.5* | ||
| At2g25270 | Unknown membrane protein | 529EALPEFSESKEIVR | 2.5* |
| At2g35350 | Protein phosphatase 2C | 175GATSGPLDPPAGEISR | 2.5–3.5 |
| At2g47000 | MDR4/PGP4 ABC transporter | 637MSSIESFKQSSLR (2P) | 2.5* |
| At3g05200 | Putative ubiquitin E3 ligase ATL6 | 401NASFLWR | 4–7* |
| 322TNSLLVLPR | 4 | ||
| At3g08510 | Phosphoinositide-specific phospholipase C | 277EVPSFIQR | 1.5–2.8 |
| At5g47910 | Respiratory burst oxidase RbohD | 341ILSQMLSQK | 16 to >20 |
Included are all phosphopeptides that were significantly induced (P= 0.05 except where indicated) in at least two biological experiments. Bold underlined letters indicate phosphorylation sites located with certainty; non-bold underlined residues are potential sites (i.e. one of the two or three sites is correct). The total number of phosphorylation sites is indicated for peptides where one site could not be localized. Complete data for each protein are listed in Supplementary Table S2.
Induction significant only at P = 0.1 in one or more experiments.
Figure 2.
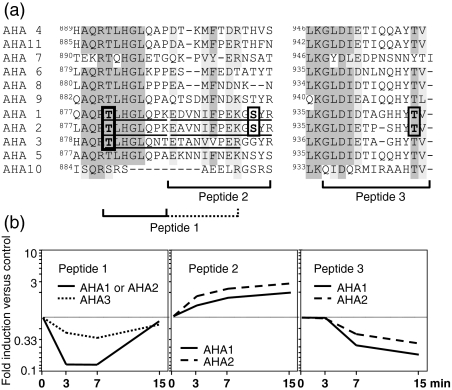
Differential regulation of three distinct phosphorylation sites on H+-ATPases. (a) ClustalW alignment of the C-terminal protein sequences of all Arabidopsis H+-ATPases. Identified phosphopeptides are marked below and their sequences are underlined within the alignment. The dashed line for peptide 1 refers to the peptide from AHA3, which is longer than peptide 1 of AHA1 and AHA2 because of the absence of an additional tryptic cleavage site. Phosphorylated residues are indicated by black frames. (b) Time course of the relative abundance (elicited versus control) of the three phosphopeptides following in vivo elicitation with flg22.
PEN3/PDR8 is a plasma membrane-resident ATP binding cassette (ABC) transporter of the pleiotropic drug resistance (PDR) family that is highly abundant both in cell culture and most plant tissues (Genevestigator data, http://www.genevestigator.ethz.ch/at/). In previous phosphoproteomic experiments (Nühse et al., 2003b and unpublished data), we identified a large number of phosphorylation sites on the N-terminus and unconserved linker domains of this protein. Here, we obtained quantitative information on the N-terminal phosphorylation sites, showing a strong increase in phosphorylation on S40, S45 and possibly one additional serine residue in response to elicitor treatment (Table 1). Another site in the central linker domain showed a more moderate induction. PEN3/PDR8 was recently identified in a genetic screen for reduced resistance to invasion by inappropriate pathogens (Stein et al., 2006). The protein accumulates at sites of attempted penetration by the non-host powdery mildew Blumeria. graminis hordei (Stein et al., 2006), and it is possible that it exports toxic compounds that limit the growth of invading pathogens. Given that our experimental system involved treatment with a bacterial peptide elicitor in a non-polarized response, we were intrigued to find induced phosphorylation sites on a protein involved in the specific polarized defence against fungi. PEN3/PDR8 may play a broader role than anticipated in basal defence and/or represent a convergence point of defence signalling against bacteria and fungi. Regulation of ABC transporters by phosphorylation, typically on the linker domains, is a widespread mechanism. The CFTR protein is one of the best-studied cases where the inhibitory effect of the linker domain is relieved by phosphorylation (Gadsby et al., 2006). An obvious hypothesis, therefore, is that the identified N-terminal phosphorylation sites activate the protein and/or determine its distribution in the membrane.
Two phosphorylation sites in the linker region of another ABC transporter, PGP4/MDR4, were also induced. This protein has been shown to regulate auxin flux in roots (Terasaka et al., 2005). A possible link between basal defence signalling and repression of auxin signalling has been discovered recently (Navarro et al., 2006), although whether MDR4 plays a role in this link remains to be shown.
In addition to identifying known components of plant defence responses, several elicitor-regulated phosphorylation sites were found in proteins that have not previously been described in the context of plant–pathogen interactions (Table 1). Elicitor-induced phosphorylation of a phosphoinositide phospholipase C (At3g08510) and dynamin (At1g59610) may reflect regulation of phospholipid signalling and/or vesicle trafficking in defence responses. The membrane protein At2g25270 has a low similarity to Drosophila Tweety ion channels based on PSI-BLAST analysis, but no known function in plants. A particularly intriguing induced phosphoprotein is the membrane-bound RING-H2 protein, ATL6. This protein is one of a large family of C3H2C3-type zinc finger proteins with an N-terminal transmembrane domain (Serrano et al., 2006), and has ubiquitin ligase activity in vitro (Stone et al., 2005). Protein ubiquitination has emerged as an important regulatory mechanism in plant immunity (Devoto et al., 2003). No role has yet been defined genetically for ATL6, but the transcript is induced by flagellin in seedlings (Navarro et al., 2004). Two ATL6 phosphorylation sites are strongly induced (Table 1), and another phosphopeptide from ATL6 was sixfold induced in a single experiment (Supplementary Table S2 and Supplementary Figure S2). These sites are in the unconserved C-terminal region and are unique for ATL6 and a closely related paralogue, ATL31 (At5g27420). While phosphorylation of the substrates of E3 ligases has been shown to be involved in the regulation of protein ubiquitination, there are relatively few examples of regulation of the E3 ligases themselves by phosphorylation (Gao and Karin, 2005). Mdm2 is a RING finger E3 ligase that targets the tumour suppressor protein p53 for degradation. Phosphorylation of Mdm2 by PKB/Akt stabilizes it by decreasing its auto-ubiquitination activity, which in turn leads to increased degradation of p53 (Feng et al., 2004). Similarly, multi-site regulation of ATL6 may either influence its activity directly or its interaction with other proteins.
Resolving complex multi-site regulation of proteins
Many of the proteins in our plasma membrane preparation contained multiple phosphorylation sites (Supplementary Table S1 and Nühse et al., 2004), a phenomenon almost certainly underestimated because of the sensitivity limits of proteomic techniques. Many of these sites may not be directly involved in a particular pathway of interest, emphasizing the need for quantification of individual phosphorylation sites to understand protein regulation. In this study, we have been able to quantify several distinct phosphopeptides for a family of H+-ATPases and found strikingly divergent regulation.
The yeast H+-ATPase Pma1 is known to be regulated at multiple phosphorylation sites during maturation in the secretory pathway and in response to glucose levels (Chang and Slayman, 1991). Little is know about the phosphoregulation of plant H+-ATPases with the exception of phosphorylation of a conserved penultimate threonine, which allows binding of a 14-3-3 protein to the C-terminus and leads to activation of the protein (Olsson et al., 1998). In an earlier paper, we reported two new phosphorylation sites at the C-terminus of different isoforms of Arabidopsis H+-ATPases (Nühse et al., 2003b). Another new phosphorylation site was discovered in this study, corresponding to T881 in either the AHA1 or AHA2 isoforms, and T882 in AHA3. The abundance of phosphopeptides including these new sites transiently decreased in response to flg22 elicitation (Figure 2b, peptide 1). In two other phosphopeptides, one or more amino acid substitutions distinguished unequivocally between AHA1 and AHA2. Phosphorylation of the penultimate threonine residue (peptide 3) was consistently reduced to one-third or less of control levels in both AHA1 and AHA2. Finally, phosphorylation of S899, found independently in AHA1 and AHA2 (peptide 2), increased threefold in both isoforms. The nearly identical dynamics of each of these sites in different paralogues indicates coordinated regulation of overall proton pumping activity. The observations here are consistent with the proposed regulation of H+-ATPase activity during response to microbial elicitors. The perception of flagellin, like all elicitors, leads to a rapid alkalinization of the growth medium, most likely through proton influx into the cell (Felix et al., 1999). Pharmacological experiments have shown that deactivation of H+-ATPases could theoretically account for most or all of this influx (Schaller and Oecking, 1999). Phosphorylation of the penultimate threonine residue is considered the major switch that releases repression of ATPase activity through the C-terminus (Palmgren, 2001). Therefore, dephosphorylation of this residue (Figure 2, peptide 3) should repress proton pumping, in agreement with the experimental findings. Whether the other phosphorylation sites with strikingly different dynamics also regulate proton pumping activity directly, or whether they affect protein stability, localization or other aspects, remains to be established.
Phosphorylation of S343 and S347 is required for the activation of RbohD but is not sufficient
RbohD is the major NADPH oxidase expressed in differentiated plant tissue as well as in cell culture. The role of ROS produced by RbohD and its partially redundant paralogue, RbohF, in defence responses is complex. While the loss of virtually all measurable superoxide production in an rbohD mutant does not affect pathogen growth or resistance in an incompatible interaction, cell death and the hypersensitive response are reduced or enhanced, depending on the pathogen (Torres et al., 2002). Later Torres et al. showed that Rboh-generated ROS control the spread of salicylate-dependent cell death surrounding infection sites (Torres et al., 2005), which suggests a role for the NADPH oxidases in signalling rather than a direct antimicrobial or cell death-inducing activity. How these enzymes are regulated in plants is poorly understood. We had previously identified up to six in vivo phosphorylation sites on RbohD (T.S.N. and S.C.P., unpublished results), of which we could now quantify three sites on two tryptic peptides. One peptide containing two sites (S343 and S347) was among the most strongly induced by the elicitor (Figure 3). These sites are unusual in that they are conserved among several of the 10 members of the Rboh protein family. We found previously that most of the phosphorylation sites identified in a large-scale qualitative study were unique to individual isoforms (Nühse et al., 2004).
Figure 3.
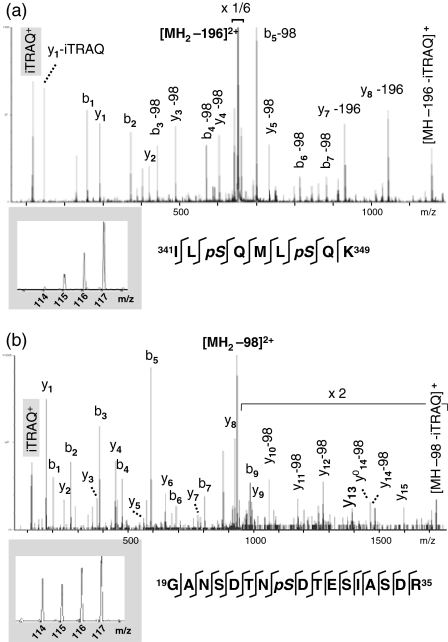
MS/MS spectra of two phosphopeptides from RbohD, covering the phosphorylation sites S343/S347 (a) and S26 (b), respectively. Below are magnifications of the m/z regions that show the iTRAQ signature ions. Peak areas at m/z 114, 115, 116 and 117 reflect the abundance of the peptide in the control, and in the 3, 7 or 15 min elicited samples, respectively.
In addition to the two strongly induced phosphorylation sites, S343 and S347 (Table 1 and Figure 3a), we quantified another RbohD phosphopeptide containing S26 (Figure 3b). The abundance of this phosphopeptide did not significantly change. The fact that S343 and S347, but not S26, are induced by the elicitor suggests that only the former two sites have a regulatory function during this response. We directly tested this hypothesis using rbohD mutant plants (Torres et al., 2002) and an in vivo luminescent assay for superoxide generation.
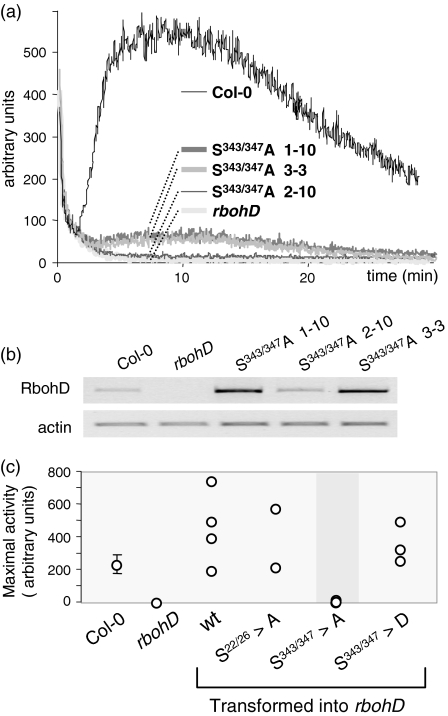
We mutagenized either S343 and S347, or S22 and S26, in genomic clones of RbohD and transformed the mutated genes under their own promoter into an Ac/Ds insertion line (rbohD) that is null for the protein (Torres et al., 2002). S22 was mutated together with S26 because a peptide containing both residues phosphorylated was frequently found in qualitative experiments, and because the duplicated NSD motif suggests phosphorylation by the same kinase. Leaf strips from the Col-0 wild-type respond to 1 μm flg22 with a strong transient oxidative burst, starting after an approximate 2 min lag phase and reaching its maximum after about 10 min (Figure 4a). The rbohD knockout mutant generates a background level of luminescence that is indistinguishable from untreated leaf tissue. Of the three independent lines of RbohD S343/347A mutants transformed into rbohD, one line (S343/347A 2-10) expressed transcript at a level comparable to the wild-type (Figure 4b), and elicitor-induced NADPH oxidase activity was undetectable in this line (Figure 4a). In two other lines with RbohD transcript expression levels much higher than in wild-type (lines 1-10 and 3-3; Figure 4b), elicitor-induced activity can be detected but is about one order of magnitude lower than in the wild-type (Figure 4a). In a separate experiment, maximal ROS-generated luminescence was recorded for several other transformants (Figure 4c). A range of activities was recorded for lines that had the wild-type RbohD gene re-transformed into the rbohD knockout. In all cases, the amount of ROS correlated with the expression level of the transcript (data not shown). Although the three S343/347A mutant lines again produced little or no ROS, neither transgenic lines expressing a S22/26A mutant were impaired in elicitor-induced superoxide production (Figure 4c), suggesting that these sites do not participate in phosphoregulation of the protein in this context. Therefore, only the differentially phosphorylated residues are required for enzyme activity.
Figure 4.
S343/347 are necessary but not sufficient for full activation of RbohD. (a) flg22-triggered oxidative burst measured by chemiluminescence in Col-0 wild-type, rbohD mutant and three independent lines expressing RbohD (S343/347A) in the rbohD background. (b) Expression level of the rbohD gene in wild-type and transformants by RT-PCR. (c) Maximal RbohD activity in Col-0 wild-type and transformants, measured as in (a). Measurements for the S343/347A mutants (three lines as above) are highlighted. Error bars for Col-0 indicate the standard error of the mean for a triplicate measurement using plants of the same age.
Interestingly, phosphorylation of S343/347 appears to be necessary but not sufficient for full activation of RbohD. The amount of ROS produced in plants expressing the phosphomimetic mutation S343/347D in the rbohD background was similar to wild-type (Figure 4c), and these plants did not constitutively produce ROS or display phenotypic symptoms (data not shown). Given the damage that uncontrolled superoxide production would do to the cell, it is perhaps unsurprising that the enzyme is under tight control from multiple regulatory mechanisms. However, what these mechanisms are is still largely an open question in plants. Like their animal homologues, plant NADPH oxidases are regulated by small G proteins (Kawasaki et al., 1999), but clear homologues of the regulatory proteins for mammalian NADPH oxidases (p40, p47 and p67) appear to be absent from plant genomes. RbohD contains a calcium-binding EF hand (Keller et al., 1998) and is calcium-activated in vitro. In recent years, non-phagocytic NADPH oxidases (Nox) have been characterized that show a number of variations in their post-translational regulation (Sumimoto et al., 2005), including human Nox5 and Duox1/2 with four EF hands. Regulation by direct phosphorylation, however, has not been reported for any of these mammalian enzymes. Therefore, the direct phosphoregulation of RbohD may reflect a new paradigm for regulation of these unusual NADPH oxidase enzymes. The elicitor-induced phosphorylation sites S343/347 of RbohD are conserved in RbohF and the largely root-specific isoforms RbohA, C, G and I. It will be interesting to test whether the activity of RbohC in the developmental pathway of root hair initiation (Foreman et al., 2003) requires the same phosphorylation sites.
A recent paper (Benschop et al., in press) describes a highly related study – the dynamics of protein phosphorylation at the plasma membrane of Arabidopsis cells in response to microbial elicitors – but using a different quantitative strategy. They metabolically labelled cells with 15N as sole nitrogen source and compared phosphopeptides from 14N-cultured control and 15N-cultured elicitor-treated cells or vice versa. In this case, the quantitative information is acquired at the MS stage rather than MS2 as with the iTRAQ labels used in the present study. Regulation of many of the flagellin-induced phosphorylation sites is in quite good agreement between the studies, including those in PEN3, the protein phosphatase 2C, ATL6, dynamin and RbohD. Our present study reports fewer phosphorylation sites as differential compared with the 15N study, partly because of differences in instrument time per experiment but largely because of the choice of statistical reference. Standard deviations for treated/control ratios based on non-phosphorylated peptides reflect variation of protein loading but not stability of phosphorylation levels. Basing significance levels on the distribution of phosphopeptides, we have discarded most changes smaller than approximately twofold. A number of peptides just below this threshold in our study show the same trend as reported by Benschop et al., as did additional phosphopeptides that we discarded because they were only found in two experiments. These results encouragingly show that in a rapidly evolving field such as proteomic technology different approaches can yield similar results.
Conclusions
Our current detailed knowledge of most signal transduction pathways is the product of many years, sometimes decades, of research into their individual constituent parts and the way they are connected. Protein phosphorylation events and activated kinases in particular have typically been identified one by one. The fruit of this painstaking labour is our ability today to integrate this knowledge into predictive models (Citri and Yarden, 2006). Recently, proteomics has joined other large-scale genomic approaches in contributing to the systems biology of signal transduction. Progress in mass spectrometric technology has revolutionized the analysis of protein-based processes. Our ability to identify protein phosphorylation sites on a large scale is one of the most striking areas of progress (Collins et al., 2005; Ficarro et al., 2002; Nühse et al., 2003b). These data have provided intriguing insight into characteristics of protein phosphorylation generally (Nühse et al., 2004), and will facilitate improvements in algorithms that predict phosphorylation sites from primary sequences (Schwartz and Gygi, 2005). Quantifying the dynamics of phosphorylation reconnects to biology, and has revealed fine regulatory details of the well-understood model system of Erb family receptors (Blagoev et al., 2004). Here, we have demonstrated the use of quantitative phosphoproteomics via iTRAQ labelling for pathway discovery and structure–function studies of individual proteins. Compared with forward genetics, this approach has the advantage of not only revealing the identity of candidates but also indicating likely regulatory sites that are relevant in this context, as we have shown for RbohD. Therefore, phosphoproteomics is an emerging strategy that is complementary to the genetic approaches prevalent in Arabidopsis research, and will greatly enhance our understanding of plant signalling pathways.
Experimental procedures
Cell culture and plasma membrane isolation
Suspension cultures of Arabidopsis thaliana ecotype Landsberg were maintained as previously described (May and Leaver, 1993; Nühse et al., 2000). Cultures (400–800 ml) were treated 6 days after subculture with 100 nm of flg22 peptide (Felix et al., 1999). After the indicated times, cells were rapidly collected by filtration, and homogenized in 2 ml g−1 fresh weight ice-cold buffer H (250 mm sucrose, 50 mm HEPES/KOH pH 7.5, 50 mm Na4P2O7, 25 mm NaF, 5% glycerol, 0.5% polyvinyl pyrrolidone, 10 mm EDTA, 1 mm Na2MoO4, 1 mm phenyl methyl sulfonyl fluoride, 25 nm K-252a, 2 nm calyculin A). Crude microsomes were isolated by differential centrifugation (10 min at 1500 g; supernatant for 45 min at 100 000 g). Microsomal pellets were rinsed in buffer R (250 mm sucrose, 5 mm potassium phosphate pH 7.5, 6 mm KCl) and then resuspended in 8 ml buffer R. Three rounds of phase-partitioning in a 24 g system (6.0% Dextran T-500, 6.0% PEG-3350 in buffer R) were performed as described previously (Nühse et al., 2003a). Plasma membranes were harvested by fivefold dilution in buffer R plus 0.02% Brij-58, and centrifugation for 60 min at 100 000 g.
Tryptic digest, iTRAQ labelling and IMAC
The plasma membrane pellet was washed once each in 100 mm Na2CO3, 500 mm triethyl ammonium bicarbonate (TEAB) and 50 mm TEAB, respectively, by resuspending and centrifugation for 45 min at 100 000 g and 0°C. After the last wash, the pellet was resuspended in 30–40 μl 50 mm TEAB, and the protein concentration determined by BCA assay (Pierce, http://www.piercenet.com). Each sample (100 μg) was denatured, digested with 10 μg sequencing grade modified trypsin (Promega; http://www.promega.com/) and iTRAQ-labelled according to the manufacturer’s instruction (Applied Biosystems; http://www.appliedbiosystems.com/). The four separate labelling reactions were combined, 400 μl water were added, and the sample lyophilized. Peptides were redissolved in 2.5% formic acid/30% acetonitrile, cleared by centrifugation (10 min at 15 000 g, room temperature) and loaded onto a 0.4 ml ICAT cation exchange cartridge (Applied Biosystems). After a 2 ml wash with solvent A (5 mm ammonium formate pH 2.7/30% acetonitrile), peptides were eluted in four steps of 0.5 ml solvent A plus 10, 30, 50 and 100 mm ammonium formate. The eluates were lyophilized, redissolved in 200 μl of 250 mm acetic acid/30% acetonitrile, and phosphopeptides isolated with 40 μl of Phos-Select resin (Sigma-Aldrich; http://www.sigmaaldrich.com/) in spin columns according to the manufacturer’s instructions. After elution with 400 mm NH4OH/30% acetonitrile, phosphopeptides were concentrated to approximately 20 μl in a vacuum, acidified with formic acid to a final concentration of 1%, and kept at −70°C until analysed.
Mass spectrometry and quantification of phosphopeptides
Detailed procedures are given in Appendix S1. Briefly, peptides were loaded directly onto a reverse-phase capillary column and eluted into the nano-electrospray ion source of a quadrupole time-of-flight mass spectrometer (Q-ToF2, Micromass UK Ltd, http://www.waters.com). Fragment ion spectra were searched using the MASCOT search tool (Matrix Science Ltd, http://www.matrixscience.com). Quantification of the m/z 114–117 peak areas was performed using the iTracker program (Shadforth et al., 2005). To distinguish true biological induction from technical variation, significance levels at P= 0.05 and P= 0.1 were calculated from a log normal distribution of the fold-change ratios for the 7 min point in each experiment. For this calculation, we excluded data from peptides with Mascot scores under 30 (with potentially poor ion statistics and thus unreliable peak area quantification).
Generation of mutant RbohD transformants
A 6.0 kb genomic fragment of rbohD containing 1.5 kb upstream sequence was amplified from genomic DNA with the primers 5′-GCGGTACCCCCCTCTAGTTCTTGTGA-3′ and 5′-GGTGGATCCGACGTAAACGCAAGAAGAC-3′, and cloned into pCAMBIA2300. Fragments between the endogenous restriction sites EcoRI/MluI or MluI/KasI were subcloned and mutagenized to generate the S22/26 or S343/347 changes, respectively (MCLAB, http://www.mclab.com). Mutant or wild-type constructs were transformed into rbohD, and homozygous transformants were identified by segregation of kanamycin resistance. Expression levels were analysed by RT-PCR, using RNA isolated with Trizol reagent (Sigma). A 1.5 kb fragment spanning the Ac/Ds insertion site in rbohD was amplified from cDNA using the primers 5′-CGGCCATCCACGCACTCAA-3′ and 5′-AACGGTCCTGAGCTTACGTGT-3′ (27 cycles, 60°C).
RbohD activity assays
Fully grown leaves of 6-week-old plants (250 mg material combined from several plants) were cut into 4 mm2 squares with a razorblade and floated on ddH2O for 6 h. The liquid was then carefully aspirated and replaced with a solution containing 1 μm flg22, 10 μg ml−1 horseradish peroxidase (Sigma, type VI-a; diluted from a 10 mg ml−1 stock in DMSO) and 50 μm luminol (diluted from a 100 mm stock in 250 mm KOH) in ddH2O. Chemiluminescence was measured in a Photek high-resolution photon counting system (HRPCS 218, camera model 6045-2/2149-1; Photek, http://www.photek.com).
Acknowledgments
We would like to thank Evonne Waterman and Matthew Smoker for technical assistance. The rbohD insertion line was kindly supplied by Professors Jonathan Jones (Sainsbury Laboratory, Norwich, UK) and Jeff Dangl (University of North Carolina, Chapel Hill, USA). This work was supported by the Gatsby Charitable Foundation and by Biotechnology and Biological Sciences Research Council grants C17990 and C510416 (to S.C.P.).
Supplementary material
The following supplementary material is available for this article online:
Distribution of fold-induction ratios for total peptides and phosphopeptides.
Annotated MS/MS spectra for all peptides in Table 1 and the previously unpublished peptide 1 from AHA 1/2 and AHA 3 (shown in Figure 2).
Full quantitative data for all phosphopeptides from all experiments whose spectra have been visually inspected.
Full quantitative data for all phosphopeptides referred to in the main text.
Methods for phosphopeptide isolation prior to iTRAQ labelling, mass spectrometry and quantification of phosphopeptides.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Asai T. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Benschop JJ. Quantitative phospho-proteomics of early elicitor signalling in Arabidopsis. Mol. Cell Proteomics. 2007 doi: 10.1074/mcp.M600429-MCP200. in press. [DOI] [PubMed] [Google Scholar]
- Blagoev B. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- Chandra S. Role of phosphorylation in elicitation of the oxidative burst in cultured soybean cells. Proc. Natl Acad. Sci. USA. 1995;92:4120–4123. doi: 10.1073/pnas.92.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J. Cell Biol. 1991;115:289–295. doi: 10.1083/jcb.115.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A. EGF–ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- Collins MO. Proteomic analysis of in vivo phosphorylated synaptic proteins. J. Biol. Chem. 2005;280:5972–5982. doi: 10.1074/jbc.M411220200. [DOI] [PubMed] [Google Scholar]
- Devoto A. Role of ubiquitination in the regulation of plant defence against pathogens. Curr. Opin. Plant Biol. 2003;6:307–311. doi: 10.1016/s1369-5266(03)00060-8. [DOI] [PubMed] [Google Scholar]
- Felix G. Rapid changes of protein phosphorylation are involved in transduction of the elicitor signal in plant cells. Proc. Natl Acad. Sci. USA. 1991;88:8831–8834. doi: 10.1073/pnas.88.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Feng J. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J. Biol. Chem. 2004;279:35510–35517. doi: 10.1074/jbc.M404936200. [DOI] [PubMed] [Google Scholar]
- Ficarro SB. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- Foreman J, et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- Gadsby DC. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M. Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol. Cell. 2005;19:581–593. doi: 10.1016/j.molcel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L. A LRR receptor-like kinase is involved in the perception of the bacterial elicitor, flagellin, in Arabidopsis. Mol. Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Gruhler A. Stable isotope labeling of Arabidopsis thaliana cells and quantitative proteomics by mass spectrometry. Mol. Cell Proteomics. 2005;4:1697–1709. doi: 10.1074/mcp.M500190-MCP200. [DOI] [PubMed] [Google Scholar]
- Kawasaki T. The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl Acad. Sci. USA. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T. A plant homolog of the neutrophil NADPH oxidase gp91(phox) subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell. 1998;10:255–266. doi: 10.1105/tpc.10.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May MJ. Oxidative stimulation of glutathion synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 1993;103:621–627. doi: 10.1104/pp.103.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004;135:1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Nühse TS. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 2000;275:7521–7526. doi: 10.1074/jbc.275.11.7521. [DOI] [PubMed] [Google Scholar]
- Nühse TS. A plasma membrane syntaxin is phosphorylated in response to the bacterial elicitor flagellin. J. Biol. Chem. 2003a;278:45248–45254. doi: 10.1074/jbc.M307443200. [DOI] [PubMed] [Google Scholar]
- Nühse TS. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell Proteomics. 2003b;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- Nühse TS. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell. 2004;16:2394–2405. doi: 10.1105/tpc.104.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 1998;118:551–555. doi: 10.1104/pp.118.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Palmgren MG. Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:817–845. doi: 10.1146/annurev.arplant.52.1.817. [DOI] [PubMed] [Google Scholar]
- Peck SC. Early phosphorylation events in biotic stress. Curr. Opin. Plant Biol. 2003;6:334–338. doi: 10.1016/s1369-5266(03)00056-6. [DOI] [PubMed] [Google Scholar]
- Peck SC. Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell. 2001;13:1467–1475. doi: 10.1105/tpc.13.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Schaller A. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell. 1999;11:263–272. doi: 10.1105/tpc.11.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- Serrano M. The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J. Mol. Evol. 2006;62:434–445. doi: 10.1007/s00239-005-0038-y. [DOI] [PubMed] [Google Scholar]
- Shadforth IP. i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics. 2005;6:145. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumimoto H. Molecular composition and regulation of the Nox family NAD(P)H oxidases. Biochem. Biophys. Res. Commun. 2005;338:677–686. doi: 10.1016/j.bbrc.2005.08.210. [DOI] [PubMed] [Google Scholar]
- Terasaka K, et al. PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell. 2005;17:2922–2939. doi: 10.1105/tpc.105.035816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl Acad. Sci. USA. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005;37:1130–1134. doi: 10.1038/ng1639. [DOI] [PubMed] [Google Scholar]
- Zipfel C. Plants and animals: a different taste for microbes? Curr. Opin. Plant Biol. 2005;8:353–360. doi: 10.1016/j.pbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Zipfel C. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of fold-induction ratios for total peptides and phosphopeptides.
Annotated MS/MS spectra for all peptides in Table 1 and the previously unpublished peptide 1 from AHA 1/2 and AHA 3 (shown in Figure 2).
Full quantitative data for all phosphopeptides from all experiments whose spectra have been visually inspected.
Full quantitative data for all phosphopeptides referred to in the main text.
Methods for phosphopeptide isolation prior to iTRAQ labelling, mass spectrometry and quantification of phosphopeptides.