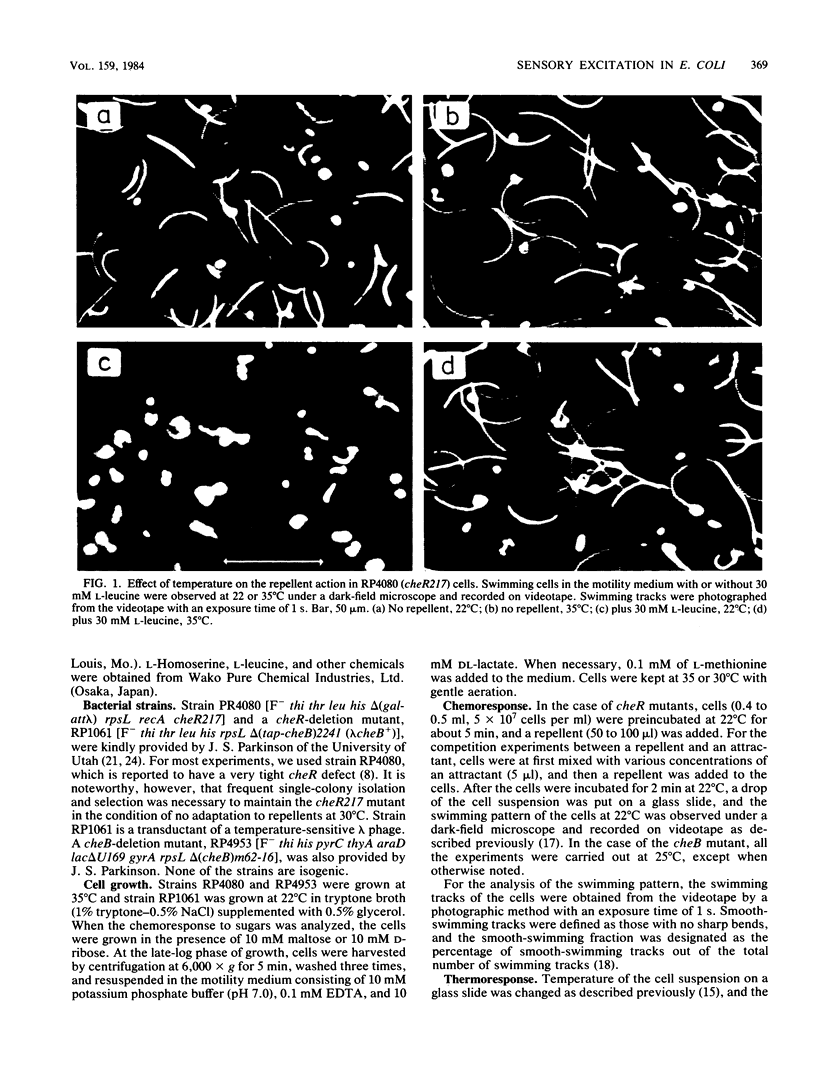
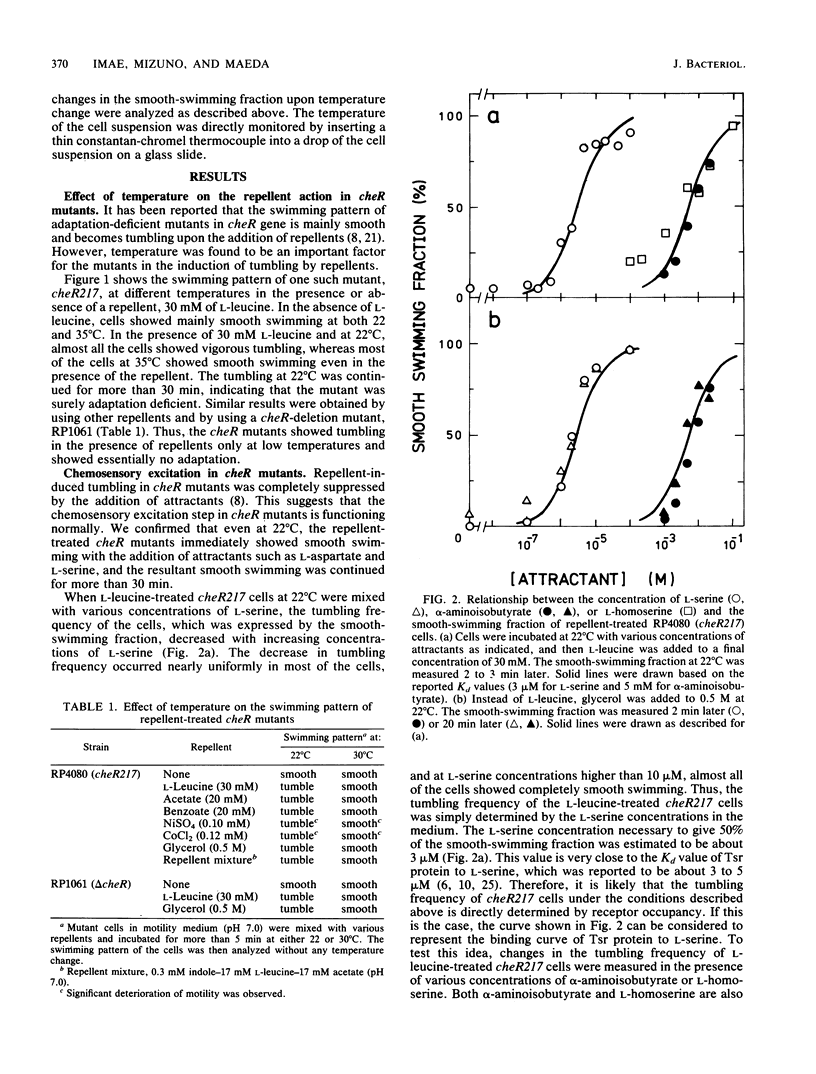
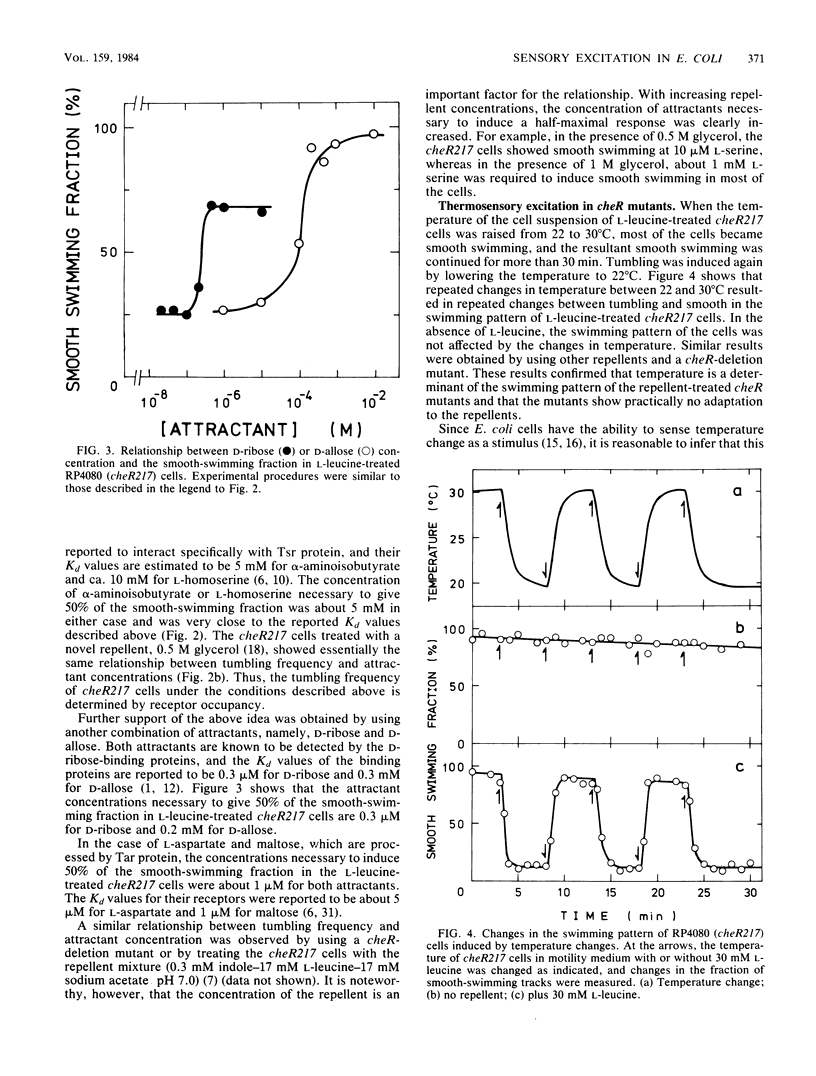
Abstract
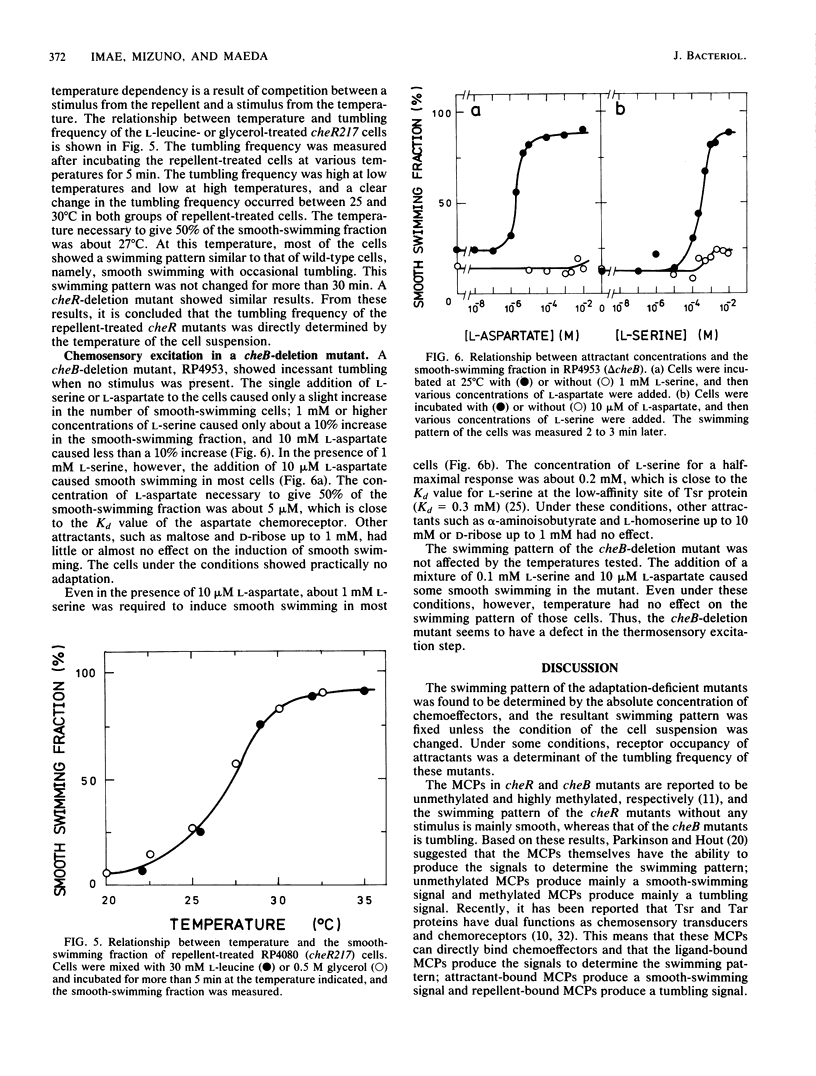
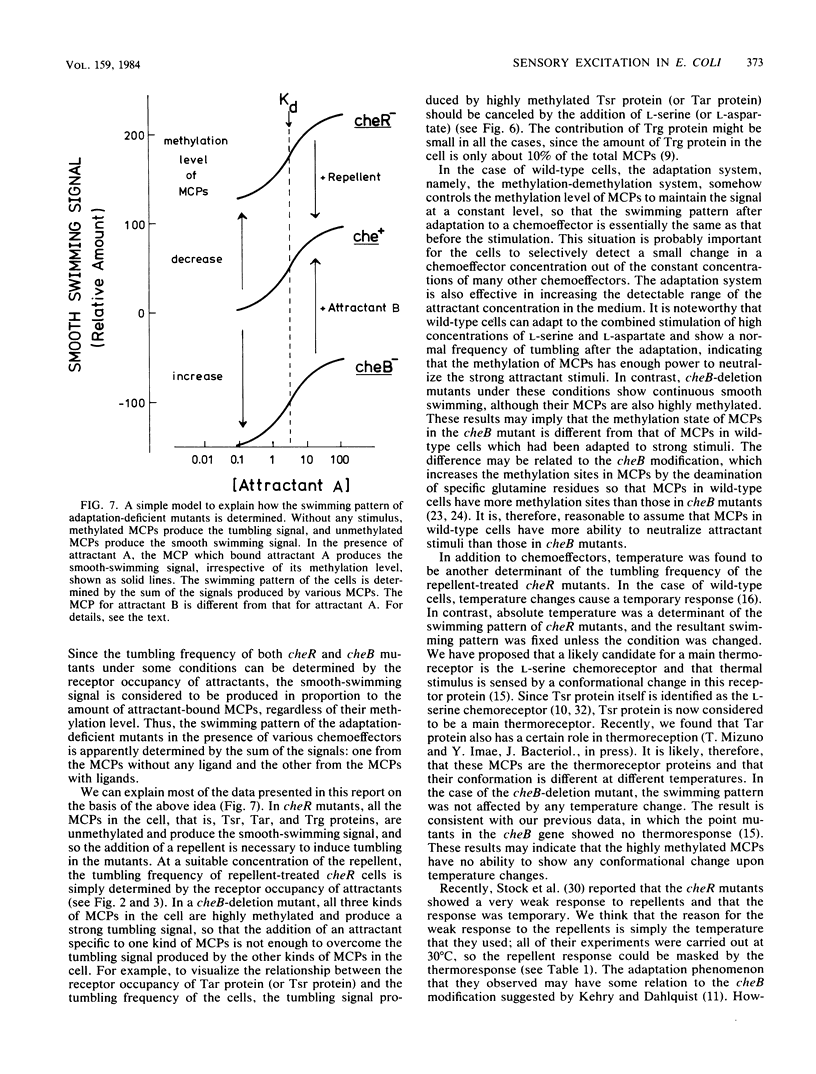
Methyl-accepting chemotaxis protein-methyltransferase-deficient mutants, cheR mutants, of Escherichia coli showed a tumble response to repellents only at low temperatures, and the resultant tumbling lasted unless the condition was changed. The swimming pattern of the repellent-treated cells was different at different temperatures, indicating that the absolute temperature is a determinant of the tumbling frequency of those cells. The tumbling of those cells was also suppressed by the addition of attractants. Under a suitable repellent concentration, the tumbling frequency of the cells was found to be simply determined by the ligand occupancy of chemoreceptors for many attractants. In a methyl-accepting chemotaxis protein-methylesterase-deficient mutant, a cheB deletion mutant, the tumbling frequency was also determined by receptor occupancy of some attractants. These results indicate that in the adaptation-deficient mutants, sensory signals are produced in proportion to the amount of ligand-bound or of thermally altered receptors and transmitted to the flagellar motors without any modification. Thus, it is concluded that the adaptation system, namely, the methylation-demethylation system of methyl-accepting chemotaxis proteins, is not concerned with the step of chemosensory or thermosensory excitation. A simple model is proposed to explain how the swimming pattern of the adaptation-deficient mutants is determined.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aksamit R. R., Koshland D. E., Jr Identification of the ribose binding protein as the receptor for ribose chemotaxis in Salmonella typhimurium. Biochemistry. 1974 Oct 22;13(22):4473–4478. doi: 10.1021/bi00719a001. [DOI] [PubMed] [Google Scholar]
- Block S. M., Segall J. E., Berg H. C. Adaptation kinetics in bacterial chemotaxis. J Bacteriol. 1983 Apr;154(1):312–323. doi: 10.1128/jb.154.1.312-323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block S. M., Segall J. E., Berg H. C. Impulse responses in bacterial chemotaxis. Cell. 1982 Nov;31(1):215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- Boyd A., Simon M. Bacterial chemotaxis. Annu Rev Physiol. 1982;44:501–517. doi: 10.1146/annurev.ph.44.030182.002441. [DOI] [PubMed] [Google Scholar]
- Brown D. A., Berg H. C. Temporal stimulation of chemotaxis in Escherichia coli. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1388–1392. doi: 10.1073/pnas.71.4.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S., Koshland D. E., Jr Membrane receptors for aspartate and serine in bacterial chemotaxis. J Biol Chem. 1979 Oct 10;254(19):9695–9702. [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Failure of sensory adaptation in bacterial mutants that are defective in a protein methylation reaction. Cell. 1978 Dec;15(4):1231–1240. doi: 10.1016/0092-8674(78)90049-1. [DOI] [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4964–4968. doi: 10.1073/pnas.74.11.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Engström P., Harayama S. Methyl-accepting chemotaxis protein III and transducer gene trg. J Bacteriol. 1981 Jan;145(1):43–49. doi: 10.1128/jb.145.1.43-49.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedblom M. L., Adler J. Genetic and biochemical properties of Escherichia coli mutants with defects in serine chemotaxis. J Bacteriol. 1980 Dec;144(3):1048–1060. doi: 10.1128/jb.144.3.1048-1060.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Dahlquist F. W. Adaptation in bacterial chemotaxis: CheB-dependent modification permits additional methylations of sensory transducer proteins. Cell. 1982 Jul;29(3):761–772. doi: 10.1016/0092-8674(82)90438-x. [DOI] [PubMed] [Google Scholar]
- Kondoh H., Ball C. B., Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R. M., Koshland D. E., Jr The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2509–2512. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K., Imae Y., Shioi J. I., Oosawa F. Effect of temperature on motility and chemotaxis of Escherichia coli. J Bacteriol. 1976 Sep;127(3):1039–1046. doi: 10.1128/jb.127.3.1039-1046.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K., Imae Y. Thermosensory transduction in Escherichia coli: inhibition of the thermoresponse by L-serine. Proc Natl Acad Sci U S A. 1979 Jan;76(1):91–95. doi: 10.1073/pnas.76.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura S., Shioi J. I., Imae Y., Iida S. Characterization of the Bacillus subtilis motile system driven by an artificially created proton motive force. J Bacteriol. 1979 Oct;140(1):28–36. doi: 10.1128/jb.140.1.28-36.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa K., Imae Y. Glycerol and ethylene glycol: members of a new class of repellents of Escherichia coli chemotaxis. J Bacteriol. 1983 Apr;154(1):104–112. doi: 10.1128/jb.154.1.104-112.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Houts S. E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982 Jul;151(1):106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Revello P. T. Sensory adaptation mutants of E. coli. Cell. 1978 Dec;15(4):1221–1230. doi: 10.1016/0092-8674(78)90048-x. [DOI] [PubMed] [Google Scholar]
- Rollins C., Dahlquist F. W. The methyl-accepting chemotaxis proteins of E. coli: a repellent-stimulated, covalent modification, distinct from methylation. Cell. 1981 Aug;25(2):333–340. doi: 10.1016/0092-8674(81)90051-9. [DOI] [PubMed] [Google Scholar]
- Sherris D., Parkinson J. S. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer W. R., Koshland D. E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A. 1977 Feb;74(2):533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Koshland D. E., Jr Quantitation of the sensory response in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Feb;72(2):710–713. doi: 10.1073/pnas.72.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Maderis A. M., Koshland D. E., Jr Bacterial chemotaxis in the absence of receptor carboxylmethylation. Cell. 1981 Nov;27(1 Pt 2):37–44. doi: 10.1016/0092-8674(81)90358-5. [DOI] [PubMed] [Google Scholar]
- Szmelcman S., Schwartz M., Silhavy T. J., Boos W. Maltose transport in Escherichia coli K12. A comparison of transport kinetics in wild-type and lambda-resistant mutants as measured by fluorescence quenching. Eur J Biochem. 1976 May 17;65(1):13–19. doi: 10.1111/j.1432-1033.1976.tb10383.x. [DOI] [PubMed] [Google Scholar]
- Wang E. A., Koshland D. E., Jr Receptor structure in the bacterial sensing system. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa H., Hayashi H., Parkinson J. S. Requirement of the cheB function for sensory adaptation in Escherichia coli. J Bacteriol. 1983 Dec;156(3):1228–1235. doi: 10.1128/jb.156.3.1228-1235.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]