Abstract
We report that human immunodeficiency virus type 1 (HIV-1) has evolved a self-perpetuating mechanism to actively generate cells permissive for productive and cytopathic infection. Only activated T cells can be productively infected, which leads to their rapid depletion (2 × 109/day in an infected individual). Establishment of productive HIV-1 infection therefore requires continual activations from the large pool of quiescent T cells. Tat protein, which is secreted by infected cells, activated uninfected quiescent T cells in vitro and in vivo. These Tat-activated uninfected cells became highly permissive for productive HIV-1 infection. Activation of primary T cells by Tat protein involved integrin receptors and was associated with activation of mitogen-activated protein kinases, including ERK1 and JNK kinase. Accordingly, these primary T cells progressed from G0 to the late G1 phase of the cell cycle.
Human immunodeficiency virus type 1 (HIV-1) infects T cells through CD4 and chemokine coreceptors (1), but infection of quiescent cells is aborted during reverse transcription, producing an unintegrated partial viral genome with a short half life (2–4). Most T cells in vivo are quiescent and therefore not permissive for efficient infection by HIV-1, so the 2 × 109 permissive T cells in an infected individual that are destroyed daily by the virus (5, 6) need to be replaced by activated T cells to sustain a productive infection. Thus, an infected individual progressively loses the quiescent T cell compartment, which is replaced by cells displaying activated phenotypes (7). Understanding this new facet of HIV-1 pathogenesis may open new opportunities for therapeutic interventions.
We hypothesize that HIV-1 encodes a priming factor that induces permissivity for infection by activating quiescent T cells to compensate for the continuous destruction of permissive cells by the cytopathic infection of HIV-1. We reported that lymphocytes treated with Tat protein exhibit signs of cell activation (8), whereas a fraction of cells undergo apoptosis, similar to treatment with polyclonal T cell activators such as anti-CD3 and phytohemagglutinin. Therefore, Tat protein could be a priming factor. Tat has the following properties that are compatible with our hypothesis: (i) it is an early viral protein and its role in HIV-1 infection extends beyond its transcriptional function (9, 10), (ii) it is released from infected cells and affects functions of uninfected cells in a paracrine fashion (8, 11–15), and (iii) it is likely to accumulate locally in lymphoid tissue where HIV-1 replication is active (16).
MATERIALS AND METHODS
Cell Cultures.
Peripheral blood mononuclear cells (PBMCs) were isolated with Ficoll–Paque (Pharmacia) (8). T cells were purified with Uni-Sorb T&B column (GIBCO/BRL). Cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum. Cells were treated with Tat for 48 h or 72 h before analysis for CD25 and Fas (CD95). In antibody blocking experiments, 12 pmol of Tat protein (12 pmol ml−1) was incubated with 5 μg of purified monoclonal antibody against Tat (Intracel) before addition to cell cultures. Alternatively, Tat was treated with 1 ml of culture supernatant of hybridoma cells containing monoclonal anti-Tat antibody or control supernatant before addition to PBMCs or T cell culture.
Immunofluorescent Staining.
Cells were washed in PBS containing 1.0% BSA and 0.1% sodium azide (wash buffer). Cells (1 × 107 cells ml−1) were incubated at 4°C in wash buffer or wash buffer containing 20 μg ml−1 anti-Fas antibody (PharMingen) for 30 min, washed, and resuspended in 2.5 μg ml−1 phycoerythrin (PE)-conjugated goat anti-mouse IgG (Coulter), and incubated at 4°C for 30 min. Cells were then washed and resuspended in PBS containing 1% paraformaldehyde and analyzed on a FACScan (Becton Dickinson). In some experiments, direct immunostaining was carried out by using 5 μg ml−1 PE-conjugated anti-Fas (PharMingen) or PE-conjugated anti-CD25 antibody 25 μg ml−1 (Coulter).
Purification of Tat Protein.
Tat protein was purified as described (17) to >95% homogeneity as determined by HPLC with a C4 reverse phase column (Vydac). The minor peak of the HPLC profile, which comprised <5% of total, was found to represent an oligomeric form of Tat according to amino acid analysis and Western blot analysis results. Tat concentration was quantitated with an Automatic Amino Acid Analyzer (Applied Biosystems). Biological activity of Tat was assayed by transactivation of the HIV-1 long terminal repeat (LTR).
Cell Cycle Analysis and cdk Kinase Assay.
Cell cycle analysis was carried out with nonapoptotic cells after staining cells with propidium iodide (8). Cell nuclear extract preparation and kinase assays were performed as described (8). For each assay, 50 μg of cell extract was immunoprecipitated with 1 μg of purified polyclonal antibody against human cyclin E. Histone H1 was used as the substrate for kinase assays.
Immunoprecipitation and Kinase Assay.
Experiments were performed essentially as described (8, 18). For each assay, 400 μg of cell extract was immunoprecipitated with 4 μg of purified goat polyclonal antibody against human Erk1 (ERK, Santa Cruz Biotechnology) or Jun N-terminal kinase (JNK; Santa Cruz Biotechnology). Purified goat IgG was used as control. Myelin basic protein was used as the substrate for mitogen-activated protein (MAP) kinase assay, and glutathione S-transferase–Jun for JNK assay.
Reverse Transcription–PCR Assay.
Total RNA was isolated by Trizol Reagent (GIBCO/BRL). The first strand of cDNA was synthesized with oligo(dT) primers and Moloney murine leukemia virus reverse transcriptase (GIBCO/BRL) (19). PCR amplification was performed with Fas primers (5′-CACTATTGCTGGAGTCATG-3′ and 5′CTGAGTCACT-AGTAATGTCC-3′) or β-actin primers (5′-CATCCTCACCCTGAAGTACC-3′ and 5′-GGTGAGGATCTTCATGAGGT-3′) for 30 cycles at 94°C for 15 sec, 60°C for 30 sec, 72°C for 1 min, and 72°C for 5 min using AmpliTaq DNA polymerase and the 9600 GeneAmp PCR system (Perkin–Elmer).
Transfection and Chloramphenicol Acetyltransferase (CAT) Assay.
U38 cells and Jurkat cells with stably transfected HIV-1 LTR CAT were treated with Tat protein for 48 h at concentrations from 12 to 1200 pmol/ml. Cells were harvested, and CAT activity was determined.
HIV-1 Infection.
Tat protein was added to PBMC or T cells after they had been cultured for 16 h. Cells were treated with Tat protein for 24 h, 48 h, 72 h, or 6 days and harvested at the indicated time points by washing three times with growth medium, and then infected with HIV-1NL4-3 for 4 h at a multiplicity of 0.5. Cells were washed with medium three times before culturing to remove residual free virus. Virus stock was prepared from supernatants of Jurkat cells transfected with pNL4-3, which encodes infectious HIV-1NL4-3 virus, and stored at −70°C. Culture supernatants were collected on days 3 and 6 after viral infection. HIV-1 p24 in culture supernatant was determined by ELISA assay (DuPont).
RESULTS
Tat Protein Activates Primary T Cells in an Antigen-Independent Way.
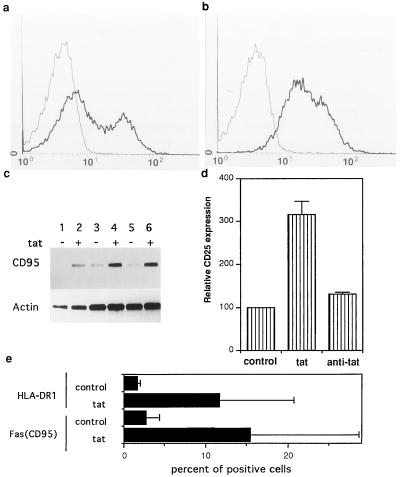
We first tested whether Tat activates lymphocytes in primary cultures of PBMCs and purified T cells isolated from healthy donors. Fas (CD95) is up-regulated in activated T cells and mediates activation-induced apoptosis (15). We determined the effect of Tat on Fas (CD95) expression by using reverse transcription–PCR and by immunofluorescent analysis. Concentrations of Tat protein used in our experiments (12–24 pmol ml−1) are close to the range of extracellular Tat demonstrated in vivo (15). Tat treatment significantly induced expression of Fas (CD95) in T cells (Fig. 1 a–c). As early as 24 h after Tat treatment, this induction was observed mainly as an increase of Fas (CD95) in cells that were negative for Fas prior to Tat treatment (Fig. 1b). The effects of extracellular Tat on PBMCs (data not shown) and purified T cells were similar, suggesting an effect primarily on the T cell population rather than on monocytes and B cells.
Figure 1.
Aberrant activation of primary T cells by Tat protein. (a and b) Fas (CD95) expression on quiescent T cells was induced by Tat protein. (a) Control T cells; (b) T cells treated with 12 pmol ml−1 Tat for 48 h. ——, Cells were stained with secondary antibody only; ——, cells were stained with anti-Fas followed by secondary antibody. T cells or PBMCs from nine blood donors were tested in independent experiments. (c) Induction of CD95 mRNA by Tat protein. Lanes 1, 3, and 5, mRNA was prepared from untreated T cells; lanes 2, 4, and 6, mRNA was prepared from Tat treated T cells. mRNA was determined by reverse transcription–PCR. From lanes 5 to 1, and, from lanes 6 to 2, cDNA was serially diluted. This is one of three independent experiments. (d) Up-regulation of CD25 expression on PBMCs by extracellular Tat. The expression of CD25 on untreated cells as measured by the mean immunofluorescent intensity was taken as 100, and CD25 expression on other samples was expressed by comparison with that of the untreated group. PBMCs were treated with Tat (12 pmol ml−1) for 48 h. T cells or PBMCs from five different donors were tested. A similar pattern of induction was observed in all experiments, although there was some variation in the absolute values in samples from different individuals. Data are mean values from three independent experiments. (e) Induction of HLA-DR1 and Fas (CD95) expression by Tat protein in mice. Data represent average of four measurements of T cells from two mice in each group. Naive T cells were purified as described, and were infused into athymic mice. After 16 h, mice were treated with Tat protein (250 μg/kg, i.v.). The control group received an equal amount of BSA in PBS. T cells were harvested from peripheral blood after 24 h.
CD25 (interleukin 2 receptor) was also used as an activation marker, because its expression correlates with T cell’s permissivity for productive infection in vivo (20). CD25 expression was determined by direct immunofluorescent staining. PBMCs displayed up-regulation of CD25 after treatment with Tat for 48 h. Purified monoclonal antibody against Tat inhibited CD25 up-regulation (Fig. 1d). Up-regulation of CD25 by 1- to 2-fold was also observed when purified T cells were treated with Tat.
To test if T cells in vivo can be activated by circulating Tat protein, we intravenously infused purified human T cells into athymic nude mice. Tat protein was injected intravenously at 250 μg/kg. As shown in Fig. 1e, Tat treatment in vivo induced expression of Fas (CD95) and also of another activation marker, HLA-DR1.
Uninfected Primary T Cells Activated by Tat Protein Are Permissive for Productive HIV-1 Infection.
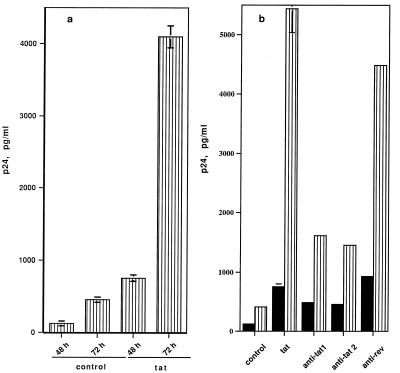
We tested whether T cells activated by extracellular Tat are permissive for productive HIV-1 infection. Quiescent T cells were activated with Tat protein before viral infection by the T cell-tropic strain, HIV-1NL4-3. As shown in Fig. 2a, Tat-activated T cells became permissive for productive HIV-1 infection as measured by p24 antigen in culture supernatants, and cells primed for 72 h were more permissive than cells treated for 48 h. This priming effect of Tat was inhibited by a monoclonal antibody against Tat or antiserum to Tat, whereas control antiserum to rev showed no significant effect (Fig. 2b). These results indicate that permissivity for viral infection was induced specifically by Tat protein. This priming effect of Tat for HIV-1 infection was also observed with PBMCs (data not shown).
Figure 2.
HIV-1 Tat primes quiescent T cells for productive HIV-1 infection (17). (a) Primary T cells were treated with Tat (12 pmol ml−1) for 48 h or 72 h before infection by HIV-1NL4-3. Levels of p24 antigen in the culture supernatant were determined on day 3 after infection. (b) Antibodies against Tat inhibit productive infection by HIV-1. A total of 24 pmol of Tat protein (12 pmol μl−1) was incubated on ice for 20 min with 10 μg of anti-Tat 1 [purified monoclonal IgG1 against HIV-1 Tat (Intracel)], or 5 μl of anti-Tat 2 (anti-serum to Tat), or 5 μl of anti-serum to rev before addition to cultures. Cells were primed with Tat protein for 48 h prior to infection. p24 levels in the culture supernatant were determined on day 3 (▪) or 6 (▥) Data in a and b represent averages of triplicates or duplicates in one of three independent experiments.
Induction of permissivity for efficient infection by Tat is due to activation of T cells rather than by transactivation of the viral LTR. First, T cells were uninfected during Tat treatment and so had no LTR. Second, the concentrations of extracellular Tat in these experiments were not sufficiently high to directly transactivate HIV-1 LTR-driven CAT expression in cells carrying this construct (data not shown). Furthermore, uninfected cells were preactivated with Tat protein for 24 h to 6 days, and then Tat was washed away from the culture before HIV-1 infection experiments.
Activation of Primary T Cells by Tat Protein Involves Integrin Receptors.
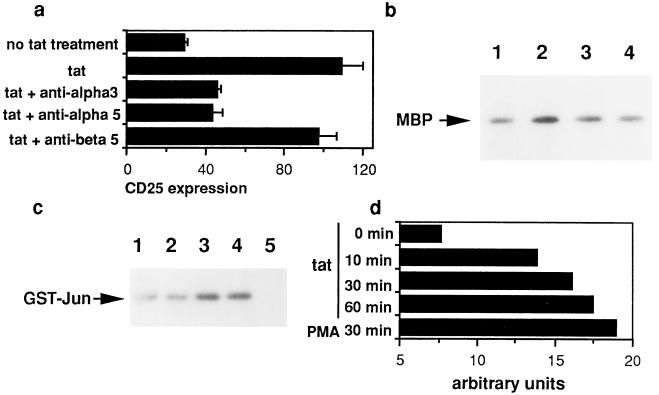
We investigated the mechanism underlying Tat-induced activation of lymphocytes starting with the hypothesis that Tat delivers an activation signal to uninfected T cells. Two signals are generally required to fully activate T cells: one is delivered through the T cell receptor, whereas the other is delivered by accessory molecule-mediated costimulatory signals. To test if Tat delivers signals through the T cell receptor complex, we determined changes in p56lck after Tat treatment. No change in phosphorylation of p56lck was detected (data not shown). Another possibility is that Tat may employ integrins as its receptor because the basic region of Tat contains an arginine-glycine-aspartic acid (RGD) motif (21). These integrin receptors have been implicated in the proliferation promoting effect of Tat on Kaposi sarcoma cells (22). Our experiments showed that monoclonal antibodies against integrin α3 and α5 inhibited Tat-induced up-regulation of CD25 (Fig. 3a), whereas an antibody to β5 showed no effect. This result was unexpected because α3 integrin is only present in moderate quantity on quiescent T cells (23). However, this result has been confirmed by four independent experiments performed on four blood donors’ cells. This result suggests a role of Tat protein at the cell surface in activating T cells. The role of accessory molecules in mediating Tat action must be complicated since several accessory pathways, such as B7-CD28, are present in T cells. Our preliminary data suggest involvement of the CD28 pathway (unpublished data).
Figure 3.
Activation of T cells by Tat protein is inhibited by antibodies against integrins (a) and is associated with activation of MAP kinase (b). (a) T cells were incubated with monoclonal antibodies to integrins β5, α3, or α5 for 1 h at 37°C. Tat protein (12 pmol/ml) was added. After 48 h, expression of CD25 was assayed. Data represent average of triplicates in one of four independent experiments. (b) Primary T cells treated with Tat protein displayed activation of MAP kinase. Lanes: 1, control T cells; 2–4, cells were treated with Tat (24 pmol ml−1) for 2 h (lane 2), 8 h (lane 3), and 24 h (lane 4), respectively. (c and d) Activation of JNK kinase (c) and MAP kinase (d) by Tat protein in Jurkat cells. Lanes: 1, control cells; 2–4, cells were treated with Tat protein at 12, 24, and 48 pmol ml−1, respectively, for 2 h; 5, control IgG for JNK antibody. GST-Jun, glutathione S-transferase–Jun. (d) Jurkat cells were treated with Tat protein for 0, 10, 30, or 60 min, or with phorbol 12-myristate 13-acetate (PMA) for 30 min, respectively. Arbitrary units represent quantitative analysis of myelin basic protein phosphorylation determined by an imaging densitometer (model GS-700; Bio-Rad).
Tat Protein Activates MAP Kinases in Primary T Cells.
If Tat protein indeed delivers an activation signal to T cells, we would expect activation of signal-transducing kinases by Tat protein. Several lines of evidence suggest that MAP kinases mediate the intracellular signaling of integrin activation as well as T cell activation (24). We determined the effect of Tat protein on MAP kinase activity. Like phorbol 12-myristate 13-acetate, Tat stimulated MAP kinase (ERK1) both in primary T cells (Fig. 3b), PBMC (data not shown), and Jurkat cells (Fig. 3d), strongly suggesting that extracellular Tat delivers an activation signal to T cells. Stimulation of MAP kinase was observed as early as 10 min after Tat treatment of Jurkat cells (Fig. 3d). Another member of the MAP kinases, JNK kinase, mediates T cell activation as well as apoptosis (25, 26). As shown in Fig. 3c, Tat protein induced JNK kinase activity. The simultaneous activation of JNK kinase and ERK1 suggests that Tat activates an upstream kinase in the MAP kinase signaling cascade. Such upstream kinases have only recently been identified (27).
Induction of G0 to G1 Phase Transition by Tat Protein in Quiescent T Cells.
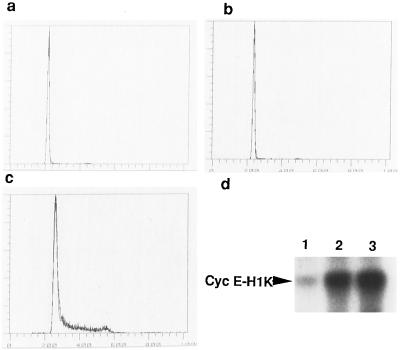
Because Tat protein activates MAP kinases, we determined cell cycle progression in naive T cells after Tat treatment. Tat alone was not sufficient to induce T cell proliferation, as measured by DNA content after 72-h treatment (Fig. 4b) and DNA synthesis determined by [3H]thymidine incorporation assay (data not shown). Failure to induce T cell proliferation by Tat alone is consistent with the “two-signal” model for T cell activation (28). Cyclin E-associated cdk activity of Tat-treated lymphocytes was induced, similar to that of proliferating lymphocytes treated with phytohemagglutinin plus human recombinant interleukin 2 (Fig. 4d), indicating that Tat drives cells to the late G1 phase of the cell cycle. This observation suggests that MAP kinase-mediated G1 cellular events are sufficient to support efficient infection by HIV-1.
Figure 4.
Aberrant activation of lymphocytes by extracellular Tat is associated with G0 to G1 progression but not entry into S phase. (a) Primary lymphocytes remained quiescent after 72 h in culture. (b) Cells activated by extracellular Tat did not enter S phase, unlike (c) cells activated by phytohemagglutinin (PHA-C; Boehringer Mannheim). (d) Primary lymphocytes treated with Tat protein for 72 h displayed activation of cyclin E associated cdk2, a late G1 kinase. Lanes: 1, control cells; 2, cells treated with Tat (24 pmol ml−1); 3, cells treated with PHA (20 μg ml−1). In a–c, PBMCs were treated with Tat protein (24 pmol ml−1), or PHA (20 μg ml−1) plus 20 units ml−1 recombinant human interleukin 2 (rIL-2; Boehringer Mannheim) for 72 h. Similar results were observed in purified T cells. In d, PBMCs were similarly treated as in b and c.
DISCUSSION
Here we report that HIV-1 Tat protein activates uninfected naive T cells and renders them permissive for productive HIV-1 infection. This nonspecific activation of immune cells by Tat protein is independent of antigen stimulation. These results suggest that HIV-1 has evolved a self-perpetuating mechanism to actively generate cells permissive for productive and cytopathic infection. This mechanism provides a reliable way for HIV-1 to compensate for the rapid destruction of activated permissive lymphocytes during the highly cytopathic infection. The induction of permissivity for viral infection is accompanied by MAP kinase activation and G1 phase progression, suggesting that G1 cellular events mediated by MAP kinase are sufficient to support productive infection by HIV-1.
Our results suggest that Tat contributes to CD4 T cell loss by two mechanisms: recruitment of quiescent T cells into a reservior that is permissive for productive HIV-1 infection and destruction by the virus, and the induction of apoptosis in uninfected cells.
After this work was completed, Ott et al. (29) reported that intracellular expression of Tat or its second exon in Jurkat cells up-regulates IL-2 secretion in response to anti-CD3 and anti-CD28 stimulations, which is consistent with our hypothesis on the importance of Tat protein in T cell pathology in HIV-1 infection (8).
The in vivo relevance of our hypothesis is supported by our preliminary in vivo experiment and further by several lines of indirect evidence. Lymphocytes in HIV-1-infected individuals are chronically and aberrantly activated (16), and Fas (CD95)-positive cells are increased (30). The relevance of Tat to these in vivo pathological changes is supported by the level of Tat achievable locally in vivo (15), and by the recent demonstration of an inverse correlation between p24 antigenemia and anti-Tat titers in HIV-infected individuals at different stages of disease progression (31). Our results suggest potential therapeutic strategies for HIV-1 infection by inhibiting extracellular Tat with neutralizing antibody or drugs, or interfering with Tat-induced aberrant cell activation events. Tat protein may serve as a target for potential AIDS vaccine development (10).
Acknowledgments
We dedicate this work to Prof. Ruth Sager with greatest respect and admiration not only for her scientific achievements but also for being a most courageous and dedicated scientist. We thank D. Gabuzda for help with HIV-1 replication studies; M. E. Hemler for helpful discussion and antibodies to integrins; S. Kharbanda for glutathione S-transferase–Jun protein; the AIDS Research and Reference Program, Division of AIDS, National Institutes of Health, for pNL4–3 (donated by M. Martin), Tat antiserum (donated by B. Cullen), rev antiserum (donated by D. Rekosh). C.J.L. was a Merck Research Scholar at the Harvard–Massachusetts Institute of Technology Division of Health Science and Technology. This work was supported by National Institutes of Health Grant AI-35576.
ABBREVIATIONS
- PBMCs
peripheral blood mononuclear cells
- JNK
Jun N-terminal kinase
- MAP
mitogen-activated protein
- CAT
chloramphenicol acetyltransferase
References
- 1.D’Souza M P, Harden V A. Nat Med. 1996;2:1293–1300. doi: 10.1038/nm1296-1293. [DOI] [PubMed] [Google Scholar]
- 2.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 3.Tang S, Patterson B, Levy J A. J Virol. 1995;69:5659–5665. doi: 10.1128/jvi.69.9.5659-5665.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCune J M. Cell. 1995;82:183–188. doi: 10.1016/0092-8674(95)90305-4. [DOI] [PubMed] [Google Scholar]
- 5.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Nature (London) 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 6.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Nature (London) 1995;373:118–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 7.Roederer M. Nat Med. 1995;1:621–622. doi: 10.1038/nm0795-621. [DOI] [PubMed] [Google Scholar]
- 8.Li C J, Friedman D J, Wang C, Metelev V, Pardee A B. Science. 1995;268:429–431. doi: 10.1126/science.7716549. [DOI] [PubMed] [Google Scholar]
- 9.Huang L-M, Joshi A, Willey R, Orenstein J, Jeang K-T. EMBO J. 1994;13:2886–2896. doi: 10.1002/j.1460-2075.1994.tb06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein G. Nat Med. 1996;1:960–964. doi: 10.1038/nm0996-960. [DOI] [PubMed] [Google Scholar]
- 11.Frankel A D, Pabo C O. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 12.Ensoli B, Gendelman R, Markham P, Fiorelli V, Colombini S, Raffeld M, Cafaro A, Chang H K, Brady J N, Gallo R C. Nature (London) 1994;371:674–680. doi: 10.1038/371674a0. [DOI] [PubMed] [Google Scholar]
- 13.Zauli G, Gibellini D, Celeghini C, Mischiati C, Bassini A, La Placa M, Capitani S. J Immunol. 1996;157:2216–2224. [PubMed] [Google Scholar]
- 14.Buonaguro L, Barillari G, Chang H K, Bohan C A, Kao V, Morgan R, Gallo R C, Ensoli B. J Virol. 1992;66:7159–7167. doi: 10.1128/jvi.66.12.7159-7167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westendorp M O, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debatin K M, Krammer P H. Nature (London) 1995;375:497–500. doi: 10.1038/375497a0. [DOI] [PubMed] [Google Scholar]
- 16.Fauci A S. Science. 1993;262:1011–1018. doi: 10.1126/science.8235617. [DOI] [PubMed] [Google Scholar]
- 17.Gentz R, Chen C-H, Rosen C A. Proc Natl Acad Sci USA. 1989;86:821–824. doi: 10.1073/pnas.86.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas S M, Demarco M, D’Arcangelo G, Halegoua S, Brugge J S. Cell. 1992;68:1031–1040. doi: 10.1016/0092-8674(92)90075-n. [DOI] [PubMed] [Google Scholar]
- 19.Tachibana O, Nakazawa H, Lampe J, Watanabe K, Kleihues P, Ohgaki H. Cancer Res. 1995;55:5528–5530. [PubMed] [Google Scholar]
- 20.Borvak J, Chou C S, Bell K, Van Dyke G, Zola H, Ramilo O, Vitetta E S. J Immunol. 1995;155:3196–3204. [PubMed] [Google Scholar]
- 21.Brake D A, Debouck C, Biesecker G. J Cell Biol. 1990;111:1275–1281. doi: 10.1083/jcb.111.3.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barillari G, Dendelman R, Gallo R C, Ensoli B. Proc Natl Acad Sci USA. 1993;90:7941–7945. doi: 10.1073/pnas.90.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemler M E. Annu Rev Immunol. 1990;8:365–400. doi: 10.1146/annurev.iy.08.040190.002053. [DOI] [PubMed] [Google Scholar]
- 24.Fields P E, Gajewski T F, Fitch F W. Science. 1996;271:1276–1278. doi: 10.1126/science.271.5253.1276. [DOI] [PubMed] [Google Scholar]
- 25.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 26.Verheij M, Bose R, Lin X H, Yao B, Jarvis W D, Grant S, Birrer M J, Szabo E, Zon L I, Kyriakis J M, Haimovitz-Friedman A, Fuks Z, Kolesnick R N. Nature (London) 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 27.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 28.Wingren A G, Parra E, Varga M, Kalland T, Sjogren H O, Hedlund G, Dohlsten M. Crit Rev Immunol. 1995;15:235–253. doi: 10.1615/critrevimmunol.v15.i3-4.30. [DOI] [PubMed] [Google Scholar]
- 29.Ott M, Emiliani S, Van Lint C, Herbein G, Lovett J, Chirmule N, McCloskey T, Pahwa S, Verdin E. Science. 1997;275:1481–1485. doi: 10.1126/science.275.5305.1481. [DOI] [PubMed] [Google Scholar]
- 30.Katsikis P D, Wunderlich E S, Smith C A, Herzenberg L A, Herzenberg L A. J Exp Med. 1995;181:2029–2036. doi: 10.1084/jem.181.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Re M C, Furlini G, Vignoli M, Ramazzotti E, Roderigo G, De Rosa V, Zauli G, Lolli S, Capitani S, La Placa M. J Acquir Immune Defic Syndr. 1996;10:408–416. doi: 10.1097/00042560-199512000-00003. [DOI] [PubMed] [Google Scholar]