Abstract
Macrophage binding of oxidatively damaged red blood cells (OxRBC) and apoptotic thymocytes correlates in many instances with a loss of phospholipid bilayer asymmetry, i.e., with an increase in expression of phosphatidylserine on the outer leaflet of the plasma membrane. Oxidatively modified LDL (OxLDL) can compete for the binding of these ligands to macrophages. However, the receptor(s) responsible remains to be identified. The present studies show that mouse peritoneal macrophages totally lacking scavenger receptor A (SRA) bound OxRBC just as effectively as wild-type macrophages, whereas their binding and uptake of acetyl LDL was reduced by more than 80%. Binding of apoptotic thymocytes and binding of OxLDL were also reduced, but only by 20–30%. We conclude that SRA is not involved in the recognition of phosphatidylserine-rich membranes but contributes to the binding of OxLDL and apoptotic thymocytes. The binding of OxRBC was almost totally calcium-dependent, whereas the binding of apoptotic thymocytes was not, suggesting that the mechanisms involved in their uptake by macrophages under these conditions were different.
Previous studies from this laboratory have suggested a relationship between macrophage receptors that recognize oxidatively damaged LDL (OxLDL) and macrophage receptors that recognize and phagocytose oxidatively damaged red blood cells (OxRBC) and apoptotic thymocytes (1). OxLDL was shown to be highly effective in blocking the binding and phagocytosis of OxRBC and also, although less completely, the binding of apoptotic thymocytes. The macrophage membrane proteins known to be able to bind OxLDL include scavenger receptors A (SRA)-I and A-II (2, 3), CD36 (and its mouse homologue) (4), CD68 (and its mouse homologue, macrosialin) (5, 6), and the FcγRII receptor (7). The latter, however, does not appear to play a major role in the internalization of OxLDL (1, 4), whereas the others have been shown to participate to a greater or lesser extent in the binding and also the internalization of OxLDL. Another member of the SRA family, designated MARCO and having close homology to SRA-I, has been cloned from a mouse macrophage library (8), but binding of OxLDL was not reported. Any of the macrophage receptors to which OxLDL binds could also be receptors to which OxRBCs and apoptotic cells bind, and this could explain the competitions previously observed. In addition, of course, there still may be unrecognized OxLDL-binding sites that participate in the binding of both OxLDL and OxRBC.
The possibility that SRA-I and SRA-II might be responsible for some of the binding of OxRBC was supported by the fact that several inhibitors of acetyl LDL (AcLDL) binding to macrophages also inhibited the binding of OxRBC, namely, polyinosinic acid, fucoidin, and malondialdehyde-modified bovine serum albumin (1). It also seemed to be supported by the fact that binding of OxRBC has long been known to correlate with an increase in the exposure of phosphatidylserine (PS) on the outer leaflet of the plasma membrane (9–11) and that competition studies showed that PS-rich liposomes could displace AcLDL from peritoneal macrophages (12). On the other hand, studies with the cloned SRA in transfected cells showed no direct binding of PS-rich liposomes (13). Furthermore, binding of OxRBC was not blocked by acetyl LDL even at very high concentrations (1). However, this still does not necessarily rule out some involvement of SRA in OxRBC binding. First, SRA is a large and complex receptor protein and could conceivably bind OxRBC at a domain different from that to which AcLDL binds. Second, Platt et al. (14) report that macrophages lacking SRA show a partial reduction in their ability to phagocytose apoptotic thymocytes. Uptake of apoptotic cells is also believed to occur in part by recognition of excess PS on the membrane (15). Thus, the role of SRA in the binding of OxRBC remains unsettled. An opportunity to settle this issue was presented by the recent success of Suzuki et al. (16) in generating SRA-negative mice by targeted disruption of the gene. The present studies were undertaken, then, to determine whether or not the binding and uptake of OxRBC and of apoptotic thymocytes are reduced in peritoneal macrophages from targeted mice lacking SRA.
MATERIALS AND METHODS
Materials.
CuSO4, diamide, tert-butylhydroperoxide, 25% glutaraldehyde, trypsin, ascorbic acid, phenylhydrazine, dexamethasone, EDTA, and cholesterol were all from Sigma; 51Cr-labeled sodium chromate and 125I-labeled sodium iodide were purchased from ICN; fetal bovine serum and penicillamine/streptomycin (PenStrep) were from Gemini Biological Products (Calabasas, CA); DMEM was from Life Technologies (Gaithersburg, MD); RPMI medium 1640 was from BioWhittaker; and acetic anhydride was from Aldrich. A monoclonal rat IgG2b directed against murine SRA (2F8) was a kind gift of S. Gordon and R.P. da Silva (Oxford University, U.K.).
Cells.
Mouse peritoneal macrophages were isolated by peritoneal lavage with PBS from homozygous SRA-targeted mice (strains SVJ × ICR) and wild-type control mice (strain SVJ). The mice were generated by Suzuki et al. (16) in the laboratory of T. Kodama (University of Tokyo, Japan) and generously sent to us. Successful targeting was verified in our animals by Southern blotting (see Results). Cells were plated in 24- or 48-well plates in DME containing 10% fetal calf serum. After the cells had been allowed to adhere for 4 hr the wells were washed three times with DME to remove serum and nonadherent cells. Binding assays were performed immediately.
Thymocytes were isolated from the thymus of 6-week-old wild-type mice, washed with ice-cold RPMI supplemented with 10% FCS. To induce apoptosis, thymocytes at 5 × 106 cells/ml were incubated with 1 μM dexamethasone in RPMI/10% FCS for 4–6 hr at 37°C, 5% CO2, 95% air, humidified. Apoptosis was confirmed by demonstrating DNA fragmentation (laddering on gel electrophoresis) and exclusion of eosin or trypan blue.
Red blood cells were collected from healthy human volunteers, washed with PBS three times, and used within 2 days. RBC (4% hematocrit) were oxidized with 200 μM CuSO4 and 5 mM ascorbic acid in PBS for 90 min at 37°C. After incubation, RBC were washed once with PBS/5 mM EDTA and once with PBS alone to remove EDTA. Treatment with trypsin (1 mg/ml), diamide, tert-butylhydroperoxide, glutaraldehyde (1 mM), and phenylhydrazine (0.1 and 1.0 mM) was carried out at a 10% hematocrit in PBS for 45 min on a shaker at 37°C. In some experiments native and oxidized RBC (100 μl of 20% hematocrit in PBS) were labeled with 50 μCi (1 Ci = 37 GBq) of 51Cr-labeled sodium chromate for 1 hr at 37°C and washed four times prior to binding experiments.
Lipoproteins.
Human LDL (d = 1.019–1.063) was isolated in EDTA (1 mg/ml) from fresh plasma by preparative ultracentrifugation (17). Protein was measured by the method of Lowry et al. (18). LDL at 100 μg/ml was oxidized by incubating overnight in PBS in the presence of 10 μM CuSO4. Acetylation with acetic anhydride was as described by Basu et al. (19). LDL was labeled with 125I for measurement of uptake and degradation (20). The extent of oxidation was assessed by measuring thiobarbituric acid reactive substances (21) and by determining electrophoretic mobility on a 1% agarose gel.
Cell Association and Degradation of Lipoproteins.
Resident mouse peritoneal macrophages (2 × 106 cells per well) were plated in 12-well plates in DME/10% FCS/PenStrep (medium A) and allowed to adhere for 2 hr. After several washes to remove nonadherent cells, the macrophages were cultured overnight in medium A and used the following day. Binding and degradation assays were performed in DME without serum or albumin. Briefly, macrophages were incubated with 125I-acetylated or 125I-oxidized LDL in the presence or absence of a 30-fold excess of unlabeled ligand. After the indicated times, the supernatant of each well was collected to determine degradation by measuring the TCA-soluble, non-iodide radioactivity (22, 23). Macrophages were solubilized with 0.2 M NaOH to measure cell association of the ligands and cell protein. Results are expressed as ng of [125I]LDL degraded or cell-associated per mg of cell protein. Total uptake is defined as the sum of LDL degraded during the incubation and the amount of 125I remaining associated with the cells at the end of the incubation.
Binding and Phagocytosis of Apoptotic Cells and Damaged Red Blood Cells.
Resident mouse peritoneal macrophages (0.6 × 106 cells/ml) were plated in 24- or 48-well plates in DME/10% FCS/PenStrep and allowed to adhere for 4 hr. Wells were washed with DME four times prior to incubation with RBC or thymocytes. Macrophages were incubated with RBC or thymocytes in DME (in the presence of inhibitors as described in figure legends) for 1 hr at 37°C in a humidified incubator. Nonadherent cells were removed by washing five times with DME, and the number of macrophages binding or having phagocytosed one or more RBCs or thymocytes was counted.
Statistical Analysis.
All cell binding and phagocytosis experiments were done in triplicate, and data are presented as the mean ± SD. Lipoprotein binding and degradation was done in duplicate. Data were analyzed with ANOVA.
RESULTS
Verification of the Deletion of SRA from the Targeted Mice.
Southern blotting with a 600-bp probe for SRA (kindly provided by T. Kodama), after digestion with XbaI/XhoI, detected a single 10-kb band in homozygous targeted mice, whereas the wild-type mice revealed a distinct band of 23 kb and no 10-kb band, as reported by Suzuki et al. (16). We also checked for expression of SRA protein by means of flow cytometry by using a monoclonal rat antibody against mouse SRA (mAb2F8) as primary antibody, followed by detection with fluorescein-conjugated goat–anti-rat F(ab)2 fragments. We detected no fluorescence on resident mouse peritoneal macrophages of the SRA knockout mice, either on the plasma membrane or intracellularly. Resident mouse peritoneal macrophages of the wild-type mice showed a low but readily detectable level of SRA protein on the plasma membrane and a large intracellular pool (i.e., after permeabilizing the cells with detergent) (data not shown).
Lipoprotein Uptake.
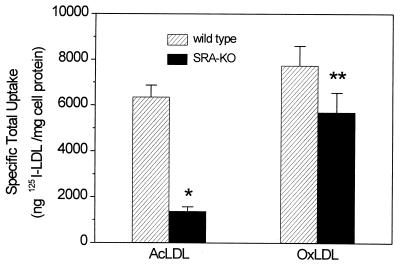
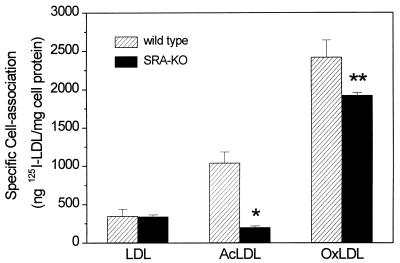
Peritoneal macrophages were incubated with 125I-labeled lipoproteins, and at the end of the incubation TCA-soluble, 125I-labeled degradation products in the medium and the 125I associated with the cells were measured. The sum of these represents the total uptake over the incubation period. Fig. 1 shows the total uptake by resident mouse peritoneal macrophages from SRA knockout and wild-type mice in the course of a 5-hr incubation at 37°C. The SRA-KO macrophages showed a highly significant, 80% reduction in total uptake of acetylated LDL. The uptake of oxidized LDL was also reduced, but only by about 30%, and this difference did not reach statistical significance. As shown in Fig. 2, binding of 125I-labeled OxLDL to the cell surface (measured at 15°C) was lower in the SRA-KO cells and the decrease was comparable to the decrease in total uptake shown in Fig. 1. Thus, the reduction in uptake of acetylated LDL and OxLDL could be accounted for as a result of a reduction in binding to the cell surface with a parallel decrease in internalization. Presumably once the ligand is bound most of it is committed to go on and be internalized and degraded.
Figure 1.
Specific uptake (sum of cell-associated 125I and 125I in degradation products in medium) of 125I-labeled acetylated and oxidized LDL by resident mouse peritoneal macrophages of wild-type (striped bars) and SRA knockout (solid bars) mice at 37°C after 5-hr incubation. ∗, P < 0.00001; ∗∗, P = 0.11. Data represent the mean of 7 experiments in duplicate ± SD.
Figure 2.
Specific cell association of 125I-labeled LDL, AcLDL, and OxLDL with wild-type and SRA knockout macrophages at 15°C. Macrophages were incubated for 2 hr with 2.5 μg 125I-labeled ligand in the presence or absence of 30-fold excess unlabeled ligand. Values shown are the mean of two determinations ± SD in a representative experiment. The experiment was done three times and gave similar results. Nonspecific binding of the label is subtracted. ∗, P = 0.019; ∗∗, P = 0.139.
Uptake of Apoptotic Cells and Damaged Red Blood Cells.
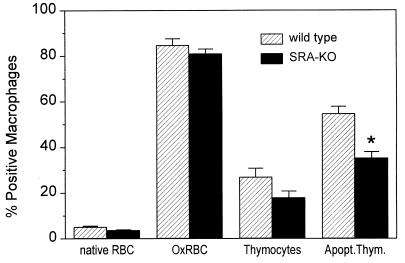
OxLDL is an effective competitor both for macrophage recognition of apoptotic cells and of OxRBC, suggesting that a common receptor may be involved (1, 24). We therefore tested whether the SRA might be participating in this type of recognition by comparing binding by wild-type and SRA-KO macrophages. RBC were oxidized by incubation with copper and ascorbic acid as described (1). This treatment probably induces a number of changes in the membrane, but the best studied of these is the exposure of PS on the outer leaflet of the membrane (9–11, 24). As shown in Fig. 3 the binding of OxRBC was almost identical in wild-type and SRA-KO macrophages, suggesting that SRA is not quantitatively important in OxRBC binding. The binding of apoptotic thymocytes, however, showed a consistent although small reduction—21% with a P value less than 0.05. In other experiments, in which we used OxRBC labeled with 51Cr, we again found no difference between wild-type and SRA-KO macrophages in the binding or phagocytosis of OxRBC. If anything, the macrophages of the SRA-KO mice showed a slight increase in binding and phagocytosis of both native and OxRBC (data not shown).
Figure 3.
Binding of RBC and thymocytes by resident mouse peritoneal macrophages of wild-type (striped bars) and SRA knockout (solid bars) mice. The data represent the mean of 17 observations in five separate experiments ± SD. ∗, P = 0.031.
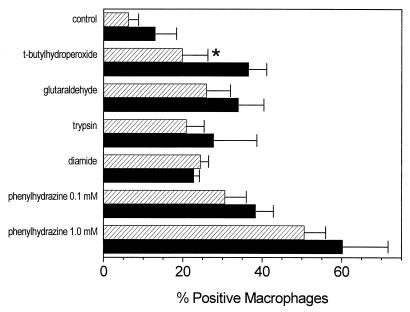
As mentioned, oxidative damage of RBC membranes may cause many changes in addition to the exposure of PS. We therefore considered it possible that although the damage induced by copper and ascorbic acid does not create a ligand recognized by the SRA, other kinds of damage might. To test this we treated RBC in a number of ways and compared binding by peritoneal macrophages from wild-type with that of macrophages from SRA-KO mice. The results are shown in Fig. 4. None of the treated RBC showed any decreased binding to macrophages from the SRA-KO mice. In the case of RBC treated with tert-butylhydroperoxide, the SRA-KO macrophages actually bound more damaged RBC.
Figure 4.
Binding of RBC by resident mouse peritoneal macrophages of wild-type (striped bars) and SRA knockout (solid bars) mice after various treatments of RBC. Data represent the mean of 4 determinations of a single experiment ± SD. ∗, P < 0.05.
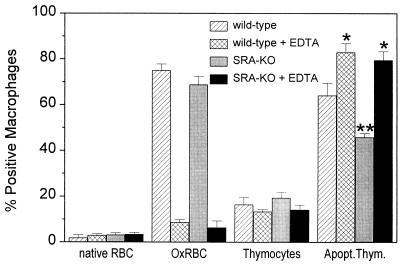
Previous results have shown that the binding of OxRBC is almost completely dependent on the presence of divalent cations (1, 24). In an attempt to further characterize the difference in recognition of oxidatively damaged RBC and apoptotic cells we investigated the effect of EDTA on the binding and uptake of apoptotic thymocytes. As shown in Fig. 5, the binding of apoptotic cells seems to be independent of divalent cations, whereas the binding of oxidized RBC is almost completely dependent on divalent cations, implying that the receptors involved are different. We have no explanation for the significant increase in binding of the apoptotic thymocytes after removal of divalent cations.
Figure 5.
The effect of 5 mM EDTA on the binding of RBC and thymocytes by macrophages of wild-type and SRA knockout mice. Data represent the mean of 3 determinations ± SD in a representative experiment. The experiment was done twice and gave the same results. ∗, P < 0.05 when compared with the values without EDTA; ∗∗, P < 0.05 for SRA-KO versus wild-type macrophages.
DISCUSSION
These studies show that most of the binding and uptake of acetyl LDL by resident mouse peritoneal macrophages is attributable to SRA, confirming the results of Suzuki et al. (16). As shown in Figs. 1 and 2, the specific binding and specific uptake of AcLDL was reduced by more than 80% in the cells from SRA-KO animals. In contrast, there was only a very small reduction in the uptake and cell-surface binding of OxLDL (approximately 30%), which did not even reach statistical significance. This finding is in agreement with results from a number of laboratories showing that only a fraction of OxLDL binding and uptake is attributable to the SRA receptor (23, 25, 26).
Under the conditions of these experiments we could see no defect in the binding and uptake of OxRBC in the macrophages lacking SRA (Fig. 3). This result is consonant with our previous finding that even high concentrations of acetyl LDL (200 μg/ml) fail to compete with OxRBC for binding to macrophages (1). Red cells damaged in other ways (treatment with glutaraldehyde, oxidation with tert-butylhydroperoxide, treatment with trypsin, diamide, or phenylhydrazine—treatments that all increase RBC binding to resident macrophages) were also bound just as effectively, if not more so, by macrophages from SRA knockout mice (Fig. 4). Finally, binding of OxRBC and apoptotic thymocytes was totally different with respect to divalent cation dependency; i.e., binding of OxRBC, as previously reported (1, 24), was almost completely inhibited by EDTA, whereas the binding of apoptotic thymocytes was, if anything, unexpectedly increased somewhat by EDTA. Recent studies using Chinese hamster ovary cells stably transfected with SRA-I or SRA-II (a gift from M. Krieger, Massachusetts Institute of Technology, Cambridge, MA) showed no binding of oxidized RBC (unpublished observation). In summary, the binding of OxRBC shows a number of striking differences from the binding of apoptotic thymocytes. If they share any receptors, those receptors must account for, at most, a small fraction of the binding of either one of these damaged cell types. We initially undertook the study of OxRBC on the assumption that this simplest cell type might be a useful model for probing the nature of the membrane changes associated with cell damage leading to recognition by macrophages. It now appears that the OxRBC may not be a suitable model, at least not for some forms of apoptotic cell recognition.
The failure of AcLDL to compete at all for the binding of OxRBC implied that SRA was not important for the binding of OxRBC. However, that finding alone did not conclusively rule it out because of two possibilities: (i) the binding of OxRBC might involve simultaneous binding to a number of SRA molecules, making the affinity of the OxRBC much higher than the affinity of acetyl LDL as a monomeric competitor; and (ii) the possibility that the binding of OxRBC might involve a larger domain on the SRA molecule, particularly the SRA-I with its large cysteine-rich domain. However, the present finding that deletion of SRA is without effect on OxRBC binding shows that SRA is not playing a significant role in OxRBC binding to mouse resident peritoneal macrophages. This is a somewhat surprising finding. One of the outstanding characteristics of SRA is its affinity for anionic compounds, including anionic lipids such as lipoteichoic acid (27) and lipid IVA from endotoxin (28). Studies by Nishikawa et al. (12) pointed to an affinity also for PS-rich liposomes, although those studies were not done with the cloned, purified receptor but rather with intact mouse peritoneal macrophages, and the conclusions were based mainly on competition studies. Lee et al. (13), using cloned receptors transfected into CHO cells, could find no evidence for binding of PS-rich liposomes. Our failure to find any decrease in binding of OxRBC in the SRA-deficient macrophage is consonant with their conclusions.
Our results, like those of Platt et al. (14), show a small but statistically significant decrease in recognition of apoptotic thymocytes in the SRA-deficient macrophage. What is the mechanism of that recognition? Apoptosis in some cases is associated with an increase in exposure of PS on the outer leaflet of the plasma membrane (15). The receptor(s) involved in this macrophage recognition has never been conclusively identified. If it were SRA, one then would expect SRA to be involved also in the binding of OxRBC. Thus, it would seem to be necessary to postulate that exposure of PS is not equivalent in the OxRBC and in the apoptotic thymocyte. Conceivably, other components of the membrane, protein or lipid, interact with PS to generate the actual receptor-binding domain.
OxLDL is also known to bind to CD36 (4), to macrosialin and its human homologue, to CD68 (5, 6), and to the FcγRII receptor (7). Which, if any of these, is involved in the binding of OxRBC remains to be determined. Recent work by Ryeom et al. (29) strongly suggests that CD36 can recognize PS-rich membranes under some conditions even though under other conditions it operates quite differently (i.e., in concert with the vitronectin receptor and thrombospondin; ref. 30). It is also possible that undiscovered receptors are involved in the joint binding of OxRBC and OxLDL.
A limitation of the present studies is that they were done using resident peritoneal macrophages only and the results do not necessarily apply to other types of macrophages. For example, there is every reason to believe that the levels of expression of various macrophage receptors will vary considerably depending on their environment. It has been shown by Fadok et al. (31) that bone marrow-derived macrophages can behave differently from resident peritoneal macrophages in the uptake of the very same population of apoptotic cells. Thus, bone marrow-derived macrophages utilized predominantly the CD36-vitronectin receptor complex for binding apoptotic thymocytes, whereas peritoneal macrophages used primarily a phosphatidyl serine-correlated recognition mechanism. Recent studies by Rose et al. (32) show that platelet activating factor and type β transforming growth factor can induce PS binding by bone marrow-derived macrophages, but the nature of the binding site is not known. The dominant mechanism or mechanisms may also vary according to the cell type of the apoptotic target and perhaps the stage in apoptosis. Finally, it becomes increasingly clear that the uptake of apoptotic cells almost certainly involves more than one mechanism.
In summary, although the binding of oxidatively damaged RBC to resident mouse peritoneal macrophages is almost completely inhibited by OxLDL, that binding does not appear to involve SRA. On the other hand, a portion of the macrophage binding of apoptotic thymocytes is attributable to SRA—approximately 20% in our studies with resident macrophages, and a higher proportion (about 50%) in the studies of Platt et al. (14) utilizing thioglycollate-elicited macrophages. The nature of the change in the cell membrane responsible for this macrophage recognition has yet to be elucidated.
Acknowledgments
We thank Drs. T. Kodama and H. Suzuki of Tokyo University for their generous gift of mice targeted for SRA. We also thank Dr. Valerie Fadok for her careful review of the manuscript. This research was supported by a Specialized Center of Research Grant on Arteriosclerosis from the National Heart, Lung, and Blood Institute (HL-14197) and by the Stein Institute for Research on Aging. V.T. was supported in part by the Dutch Heart Foundation (Nederlandse Hart Stichting).
ABBREVIATIONS
- OxRBC
oxidatively damaged human red blood cells
- OxLDL
oxidatively modified low-density lipoproteins
- SRA
scavenger receptor A
- SRA-KO
SRA knockout
- PS
phosphatidylserine
Note Added in Proof
After this manuscript was submitted, Lougheed et al. (3) reported very similar studies using SRA-KO macrophages. They found an 80% decrease in acetyl LDL uptake in the mutant cells but only a 30% decrease in uptake of oxidized LDL, findings very similar to those reported here. Binding of OxRBC was not studied.
References
- 1.Sambrano G R, Parthasarathy S, Steinberg D. Proc Natl Acad Sci USA. 1994;91:3265–3269. doi: 10.1073/pnas.91.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman M, Ekkel Y, Rohrer L, Penman M, Freedman N J, Chisolm G M, Krieger M. Proc Natl Acad Sci USA. 1991;88:4931–4935. doi: 10.1073/pnas.88.11.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lougheed M, Lum C M, Ling W, Suzuki H, Kodama T, Steinbrecher U. J Biol Chem. 1997;272:12938–12944. doi: 10.1074/jbc.272.20.12938. [DOI] [PubMed] [Google Scholar]
- 4.Endemann G, Stanton L W, Madden K S, Bryant C M, White R T, Protter A A. J Biol Chem. 1993;268:11811–11816. [PubMed] [Google Scholar]
- 5.Ottnad E, Parthasarathy S, Sambrano G R, Ramprasad M P, Quehenberger O, Kondratenko N, Green S, Steinberg D. Proc Natl Acad Sci USA. 1995;92:1391–1395. doi: 10.1073/pnas.92.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramprasad M P, Fischer W, Witztum J L, Sambrano G R, Quehenberger O, Steinberg D. Proc Natl Acad Sci USA. 1995;92:9580–9584. doi: 10.1073/pnas.92.21.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanton L W, White R T, Bryant C M, Protter A A, Endemann G. J Biol Chem. 1992;267:22446–22451. [PubMed] [Google Scholar]
- 8.Elomaa O, Kangas M, Sahlberg C, Tuukkanen J, Somunen R, Liakka A, Thesleff I, Kraal G, Tryggavason K. Cell. 1995;80:603–609. doi: 10.1016/0092-8674(95)90514-6. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y, Schroit A J. J Biol Chem. 1983;258:11335–11343. [PubMed] [Google Scholar]
- 10.Schroit A J, Madsen J W, Tanaka Y. J Biol Chem. 1985;260:5131–5138. [PubMed] [Google Scholar]
- 11.McEvoy L, Williamson P, Schlegel R A. Proc Natl Acad Sci USA. 1986;83:3311–3315. doi: 10.1073/pnas.83.10.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishikawa K, Arai H, Inoue K. J Biol Chem. 1990;265:5226–5231. [PubMed] [Google Scholar]
- 13.Lee K D, Pitas R E, Papahadjopoulos D. Biochim Biophys Acta. 1992;1111:1–6. doi: 10.1016/0005-2736(92)90267-p. [DOI] [PubMed] [Google Scholar]
- 14.Platt N, Suzuki H, Kurihara Y, Kodama T, Gordon S. Proc Natl Acad Sci USA. 1996;93:12456–12460. doi: 10.1073/pnas.93.22.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fadok V A, Voelker D R, Campbell P A, Cohen J J, Bratton D L, Henson P M. J Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 16.Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, et al. Nature (London) 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 17.Havel R J, Eder H A, Bragdon J H. J Clin Invest. 1955;34:1345–1353. doi: 10.1172/JCI103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowry O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Basu S K, Goldstein J L, Anderson G W, Brown M S. Proc Natl Acad Sci USA. 1976;73:3178–3182. doi: 10.1073/pnas.73.9.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salacinski P R P, McLean C, Sykes J E C, Clement-Jones V V, Lowry P J. Anal Biochem. 1981;117:136–146. doi: 10.1016/0003-2697(81)90703-x. [DOI] [PubMed] [Google Scholar]
- 21.Sparrow C P, Parthasarathy S, Steinberg D. J Lipid Res. 1988;29:745–753. [PubMed] [Google Scholar]
- 22.Steinbrecher U P, Parthasarathy S, Leake D S, Witztum J L, Steinberg D. Proc Natl Acad Sci USA. 1984;81:3883–3887. doi: 10.1073/pnas.81.12.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henriksen T, Mahoney E M, Steinberg D. Proc Natl Acad Sci USA. 1981;78:6499–6503. doi: 10.1073/pnas.78.10.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrano G R, Steinberg D. Proc Natl Acad Sci USA. 1995;92:1396–1400. doi: 10.1073/pnas.92.5.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arai H, Kita T, Yokode M, Narumiya S, Kawai C. Biochem Biophys Res Commun. 1989;159:1375–1382. doi: 10.1016/0006-291x(89)92262-6. [DOI] [PubMed] [Google Scholar]
- 26.Sparrow C P, Parthasarathy S, Steinberg D. J Biol Chem. 1989;264:2599–2604. [PubMed] [Google Scholar]
- 27.Dunne D W, Resnick D, Greenberg J, Krieger M, Joiner K A. Proc Natl Acad Sci USA. 1994;91:1863–1867. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hampton R Y, Golenbock D T, Penman M, Krieger M, Raetz C R. Nature (London) 1991;352:342–344. doi: 10.1038/352342a0. [DOI] [PubMed] [Google Scholar]
- 29.Ryeom S, Silverstein R, Scotto A, Sparrow J. Am Soc Biochem Mol Biol. 1996;271:20536–20539. doi: 10.1074/jbc.271.34.20536. [DOI] [PubMed] [Google Scholar]
- 30.Savill J, Fadok V, Henson P, Haslett C. Immunol Today. 1993;14:131–136. doi: 10.1016/0167-5699(93)90215-7. [DOI] [PubMed] [Google Scholar]
- 31.Fadok V A, Savill J S, Haslett C, Bratton D L, Doherty D E, Campbell P A, Henson P M. J Immunol. 1992;149:4029–4035. [PubMed] [Google Scholar]
- 32.Rose D, Fadok V, Riches D, Clay K, Henson P. J Immunol. 1995;155:5819–5825. [PubMed] [Google Scholar]