Abstract
The product of the WT1 Wilms tumor suppressor gene controls the expression of genes encoding components of the insulin-like growth factor and transforming growth factor β signaling systems. The role of these growth factors in breast tumor growth led us to investigate possible WT1 gene expression in normal and cancerous breast tissue. WT1 was detected by immunohistochemistry in the normal mammary duct and lobule, and the patterns of expression were consistent with developmental regulation. In a survey of 21 infiltrating tumors, 40% lacked immunodetectable WT1 altogether and an additional 28% were primarily WT1-negative. Cytoplasmic, but not nuclear, localization of WT1 was noted in some tumor cells and WT1 was detected, sometimes at high levels, in more-advanced estrogen-receptor-negative tumors. In this highly malignant subset, the tumor suppressor protein p53, which can physically interact with WT1, was also sometimes detected. WT1 mRNA was detected in normal and tumor tissue by reverse transcription-coupled PCR. Alternative splicing of the WT1 mRNA may regulate gene targeting of the WT1 protein through changes either in its regulatory or zinc-finger domains. The relative proportions of WT1 mRNA splice variants were altered in a random sample of breast tumors, providing evidence that different tumors may share a common WT1-related defect resulting in altered regulation of target genes.
Normal growth and differentiation of the mammary gland depend on endocrine hormones that act in concert with locally produced growth factors such as the insulin-like growth factors (IGFs) and members of the transforming growth factor β (TGF-β) family. Multiple lines of evidence support the role of IGFs, acting through the IGF-I receptor (IGF-IR), in normal mammary growth and morphogenesis and in mammary tumorigenesis (1–6). In vivo, IGF-I, supplemented with estrogen, orchestrated normal ductal growth and morphogenesis when administered adjacent to regressed mammary epithelium in the rat (7). IGFs are potent mitogens in numerous breast cancer cell lines and expression of the IGF-IR, which is found in high concentrations in primary breast cancers, is crucial for tumor cell proliferation; blockade with an IGF-IR-specific antibody inhibited IGF-stimulated cell division in vitro and tumor formation in vivo (2). The TGF-β system appears responsible for the normal inhibition of mammary growth (8–10). Paradoxically, expression of TGF-β in breast tumors is correlated with metastasis and poor prognosis, and TGF-β can stimulate the tumorigenicity of breast cancer cell lines in nude mice (11–14). The genes encoding the IGF-IR, IGF-II, and TGF-β, as well as WT1 itself, are among the targets of the product of the Wilms tumor suppressor gene WT1 (15–18), which encodes a transcription factor consisting of an amino-terminal regulatory domain and a carboxyl-terminal domain composed of four Cys2His2 zinc-finger motifs responsible for DNA and RNA binding (19, 20). An alternative splice site in each of these domains results in four isoforms of WT1 mRNA (21). Mutations in the WT1 gene are associated with a subset of Wilms tumors, the most common pediatric renal cancer (22–24). It has been previously proposed that, during normal renal development, WT1 functions to suppress an IGF-II/IGF-IR autocrine loop to effect differentiation of the renal epithelium and that loss of WT1 function contributes to Wilms tumorigenesis through constituitive activation of this loop (25).
The causative role of the loss of the WT1 transcription factor in the etiology of a human tumor and its regulation of genes encoding at least two growth factors and a tyrosine kinase known to be important in mammary duct growth regulation and breast cancer cell proliferation led us to investigate possible WT1 expression in the normal and cancerous breast. We now report, to our knowledge, the first evidence that WT1 protein is present in normal breast tissue and appears to be developmentally regulated and that a high percentage of breast tumor cells express little or no WT1 protein. WT1 mRNA was also detected, and differences in the proportions of alternatively spliced WT1 mRNAs correlated with normal versus cancerous status.
EXPERIMENTAL PROCEDURES
Tissue.
Specimens used for immunohistochemical analysis were obtained directly after surgical excision, transferred immediately to chilled (4°C) 4% paraformaldehyde in phosphate-buffered saline (PBS), and fixed for 3 h. Additional specimens of fixed sectioned breast tumors were provided by the University of Michigan Breast Cell/Tissue Bank, where histological grading, steroid hormone receptor status, and p53 and cERB2 expression were determined. Specimens used for RNA extraction were quick-frozen in liquid nitrogen immediately after excision. Histological typing of these specimens was determined by Kelly R. O’Keefe, Dominican Hospital, Santa Cruz, CA. Steroid hormone receptor status was available only for a subset of these samples.
Immunohistochemistry.
Fixed tissue was dehydrated through a graded series of ethanols to xylene and embedded in paraffin wax. Tissue was then sectioned at 7 μm and mounted on slides coated with 3-aminopropyltriethoxysilane (Sigma). The anti-WT1 antibody used in this study was WT(C-19) (sc-192; Santa Cruz Biotechnology) directed against an epitope corresponding to the 9 amino acids at the carboxyl terminus of the human WT1 protein. It was used at 1:200 dilution in PBS. A second anti-WT1 antibody, WT(180) (sc-846; Santa Cruz Biotechnology) specific for the amino terminus was used at a 1:10 dilution. Sections were incubated with antibody overnight at room temperature and antibody binding was detected with the avidin-biotin-peroxidase system protocol for the Vectastain standard kit with the following additional blocking steps: aldehyde groups were blocked using 0.2% glycine in PBS for two 5-min periods; endogenous peroxidases with 0.3% hydrogen peroxide were blocked in methanol for 30 min; nonspecific proteins were blocked with 2% dried milk in PBS for 30 min and 5% goat serum in PBS for 3 h instead of 30 min (Vector Laboratories).
Evidence for Specificity of Anti-WT1 Antibody C-19.
In COS7 cells, nuclear staining by C-19 was observed only in transfectants containing WT1-expressing constructs; transfectants expressing a mutant WT1 lacking the carboxyl-terminal domain did not stain nor did untransfected cells (26). In this same study, C-19 colocalized with each of four independently raised anti-WT1 monoclonal antibodies in COS cells as well as in mouse testis and kidney tissue (26). WT1 expressing and nonexpressing cell lines (identified by Northern blot hybridization) were tested for staining with C-19 antibodies or monoclonal antibodies, and expressing cell lines all showed characteristic nuclear staining pattern with C-19 or monoclonal antibodies, whereas nonexpressing cells were not stained (26). In separate studies, buffalo rat BRL-3A cells and human glomerular epithelial cells were shown to express WT1 and demonstrated nuclear staining with C-19 (27, 28). Additionally, Western immunoblotting of antibody C-19 gave a single band of correct size in extracts from CHO cells containing a WT1 expression vector (29). Finally, transfection of epitope-tagged WT1 into a variety of WT1-negative cell lines, followed by immunostaining and/or Western immunoblotting with C-19 and an anti-FLAG (M2) monoclonal antibody verified the specificity of C-19 in our own hands.
The specificity of WT1 immunostaining in our system is supported by the determination that a second independently raised polyclonal antibody, WT(180) (sc-846; Santa Cruz Biotechnology), directed against the amino-terminal domain of WT1 gave the same staining pattern as C-19 (data not shown), and preincubation of C-19 with its cognate peptide greatly reduced nuclear staining (Fig. 1A Inset).
Figure 1.
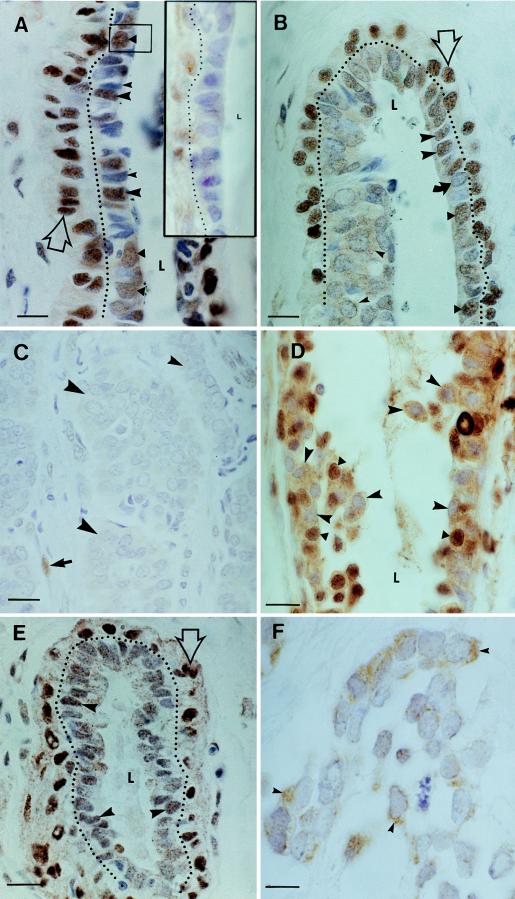
Photomicrographs illustrating immunolocalization of WT1 protein in the epithelial cells of normal and cancerous human breast tissue. Cells that are positive for WT1 protein stained brown; the blue nuclear stain is hematoxylin. (A) Nuclear morphology and WT1 staining characteristics of normal mammary ductal cells. Cells with oval nuclei and diffuse chromatin were WT1-positive (triangles and box in the center). Cells with polygonal nuclei and compact chromatin were positive (large arrowheads) or negative (small arrowhead) for WT1. The latter stained deeply with hematoxylin. Myoepithelial cells were WT1-positive (open arrow; dotted line delineates the boundary between lumenal and myoepithelial cells). L, ductal lumen. (Inset) Antibody preincubated with cognate peptide shows reduced staining. Myoepithelial cells are on the left of dotted line. (Bar = 15 μm.) (B) Ductal tip partially involved with carcinoma in situ. Normal cellular arrangement occurred on the right side of the duct; a multilayered tumor appears on left side. L, lumen. The top of the L points to transitional zone between the normal and tumorous sides of the duct. Immunostaining of monolayer of normal-appearing ductal cells on the right side of the duct: some ductal cells with oval nuclei and diffuse chromatin were WT1-positive (triangles); fewer were WT1-negative (curved arrow). Numerous cells with polygonal nuclei were WT1-positive (large arrowheads). The tumor element consists primarily of cells with large oval WT1-negative nuclei containing diffuse chromatin. Some cells in this mass had WT1-positive cytoplasm (small arrowheads). Myoepithelial layer lies outside dotted line. (Bar = 15 μm.) (C) Infiltrating ductal carcinoma. Tumor cells have proliferated to form masses (large arrowheads) within which all cells were WT1-negative. A band of infiltrating tumor cells is visible (small arrowhead). This tumor and the previously described tumor in situ are from the same patient. WT1-positive stromal cell is positive control for immunostaining. (Bar = 20 μm.) (D) Nuclear and cytoplasmic immunostaining for WT1 in infiltrating ductal carcinoma. WT1 immunostaining subdivides this population roughly in half on the basis of cytoplasmic versus nuclear localization of the protein. Cells with only cytoplasmic WT1 immunoreactivity (arrowheads) are intermixed with cells having nuclear stain. In addition, there are some cells that have both cytoplasmic and nuclear WT1 (triangles). (Bar = 20 μm.) (E) Normal mammary lobule. Normal-appearing lumenal epithelium stained with moderate intensity for WT1 (arrowheads). The nuclei of the myoepithelial cells stained deeply for WT1 (open arrow; dotted line delineates boundary between myoepithelial and lumenal cells). (Bar = 20 μm.) (F) Infiltrating lobular carcinoma. Section from vicinity of normal lobule (E). The nuclei of tumor cells are uniformly negative for WT1; however, some cells showed cytoplasmic staining (arrowheads). (Bar = 15 μm.)
RNA Preparation and Reverse Transcription Reactions.
Frozen samples were pulverized under liquid nitrogen, and a primary extraction of total RNA was performed with Purescript reagents (Gentra Systems) and was followed by secondary purification with a Qiagen total RNA midi kit (Qiagen, Chatsworth, CA). Reverse transcription reactions used an NN(T)33 primer (CLONTECH) with Moloney murine leukemia virus reverse transcriptase and a standard buffer (Promega) in a 20-μl reaction volume containing 10 μg of human breast tissue total RNA or 3 μg of human kidney total RNA. High molecular weight reaction products were purified from the reaction mixture by centrifugal filtration using a Millipore filter with a 30,000 molecular weight cut-off. Purified DNA/RNA was resuspended in 25 μl of RNase-free water for PCR. Samples of human kidney and uterus RNA were from (CLONTECH).
PCR.
One-tenth of the purified DNA/RNA volume was subjected to a first-round PCR amplification (94°C for 1 min, 58°C for 2 min, and 72°C for 3 min for 30 cycles) using Taq polymerase (Fisher Scientific) in a 50-μl reaction volume in a Perkin–Elmer model 9600 thermal cycler. For the second round, a 1:25 dilution of first-round reaction mixtures containing mammary samples or a 1:50 dilution of the kidney or uterus RNA was made in fresh reaction mixtures containing a second set of primers and the same program was repeated for an additional 25 cycles. WT1-specific PCR primer sequences were selected using macvector primer selection software (IBI-Kodak) and are as follows: F3 (20-mer), 5′-TTGTGATGGCGGACAAATTC-3′; F1 (21-mer), 5′-GGAATCAGATGAACCTAGGAG-3′; B5 (25-mer), 5′-CGTTTCTCACTGGTCTCAGATGCCG-3′; F11 (24-mer), 5′-AGGTTTTCTCGCTCAGACCAGCTC-3′; B1 (20-mer) 5′-GCCACCGACAGCTGAAGGGC-3′; B3 (20-mer), 5′-TTGTGATGGCGGACMAATTC-3′.
Gel electrophoresis in 2% agarose was used to resolve both splice variants. Low EEO agarose (Fisher Scientific) in 1× TAE (47) was used to resolve the 51-bp difference in the exon 5 variants. “Metaphor” high-resolution agarose (FMC) in 1× TBE (47) was used to resolve the 9-bp difference in the KTS variants.
WT1 Southern Blot Hybridization.
Probe was a digoxigenin-labeled 645-bp human WT1 cDNA fragment spanning exons 7 through 10 and into the 3′ untranslated region. Hybridization was at 45°C overnight followed by two 30-min washes in 0.1× standard saline citrate/0.1% SDS at 65°C. Signal was detected by chemiluminescent substrate diluted 1:1 in basic buffer (Lumi-Phos 530, Genius buffer 3; Boehringer Mannheim, Indianapolis, IN).
Determination of the Relative Proportions of Alternative Splice Variants.
Each sample of total RNA from a reduction mammoplasty or breast cancer patient was subjected to at least two reverse transcription reactions and from two to six primary (round 1) PCR amplifications prior to amplification with splice-specific primers. For each sample therefore, up to six replicate PCR amplification reactions were carried out with multiple samplings of the reverse-transcribed RNA. Within the normal or tumor groups, the data from all patients gave essentially the same results and, therefore, were pooled, and the occurrence of signals for unspliced and spliced forms, appearing either independently or together, were expressed as a percentage of the total number of occurrences observed for each form.
RESULTS
WT1 expression in normal breast tissue was investigated by immunohistochemistry with a polyclonal antibody directed against the WT1 carboxyl terminus. Normal mammary ducts are constructed of cells from two developmental lineages. The first, which always stained heavily with antibodies to WT1, is the myoepithelium, a sheath of contractile cells (Fig. 1A, open arrow) that overlays cells of the second lineage that line the duct lumen. The latter population can be further subdivided on the basis of nuclear morphology and chromatin density. Cells with rounded nuclei and diffuse chromatin were mostly WT1-positive (Fig. 1A; solid triangles). Cells with this appearance have been described in the mouse mammary gland and are considered to be less-differentiated probable stem cells that to give rise to lobular cells or new ducts (30–33). The second ductal cell type has a polygonal nucleus with compact chromatin, features that are considered characteristic of differentiated cells. Some of these cells were also positive for WT1 (Fig. 1A).
The complex expression pattern for WT1 observed in ductal cells was not seen in the normal lobule, where staining was more uniform, presumably reflecting the fact that these more-differentiated presecretory cells constitute a relatively homogeneous population (Fig. 1E). As with the duct, the myoepithelial cells investing lobules stained heavily for WT1.
In contrast to normal mammary epithelium, tumor cells often lacked detectable WT1 protein. All the cells of one ductal tumor, for example, uniformly lacked immunodetectable WT1; the nuclei of these tumor cells were monomorphic and similar to putative less-differentiated cells described for the normal duct (compare the nuclear morphology in Fig. 1C with Fig. 1A, triangle in box). The WT1-deficient phenotype can be established early in tumorigenesis, as seen in an example of carcinoma in situ, a situation where tumor elements are still contained within an otherwise normal duct (Fig. 1B). As pictured, a multilayered tumor that is negative for nuclear WT1 protein lies directly to the left of a normal-appearing duct wall containing numerous WT1-positive cells. Lobular tumors were also WT1-negative (Fig. 1F). In the pictured example, infiltrating tumor cells formed WT1-negative acinar-like structures or small islands, the larger of which had myoepithelial-like cells at their periphery that stained for WT1.
Changes in the intracellular localization of WT1 protein accompanied tumor progression. WT1 was detected in the cytoplasm of tumor cells in the carcinoma in situ but not in the infiltrating tumor from the same patient (Fig. 1 B and C, respectively). Cells of a second infiltrating ductal carcinoma had either cytoplasmic or nuclear WT1, suggesting that further differentiation of WT1 expression may have occurred after clonal transformation (Fig. 1D). Cytoplasmic staining for WT1 was also detected in a lobular carcinoma (Fig. 1F).
To determine the frequency of the WT1-negative breast tumor phenotype, levels of expression were investigated in tissues from 21 patients with infiltrating mammary carcinoma (Table 1). All histopathological grades of ductal tumor were represented and patients ranged in age from 29 to 88 years. In 40% of all tumors studied, WT1 protein was undetectable. Where tumor cytology was heterogeneous, a majority cell type (estimated by inspection to be 50% or more of the tumor cells) was usually identifiable and in 28% of these tumors, the majority of the cells were WT1-negative. A higher percentage of lobular tumors compared with ductal carcinomas (83% versus 67%) had a majority of WT1-negative cells (data not shown). A correlation was noted between estrogen receptor status and WT1 expression; 78% of the lower-grade receptor-positive tumors lacked WT1 compared with only 40% for receptor-negative counterparts (data not shown).
Table 1.
WT1 protein expression: Survey of immunostaining in normal and cancerous breast tissue
| Tissue | WT1 Neg | WT1 Pos |
|---|---|---|
| Tumor | 15 | 6 |
| Normal | 1 | 14 |
In 60% (9 of 15) of the tumors surveyed, WT1 immunostaining was absent in greater than 90% of the tumor cells (estimated by inspection). In the remaining 6 tumors, 50% or more of the tumor cells were estimated to be WT1-negative. Source of normal tissue: ducts and lobules with normal-appearing cellular architecture and nuclear morphology were often found in the vicinity of tumors and were used as controls (Normal).
The detection of the p53 tumor suppressor protein and estrogen-receptor-negative status are associated with higher-grade more-aggressive cancers that have the poorest clinical outcomes (34). Although the sample numbers were too small to assess statistical significance, the observation that two of three WT1-positive high- grade tumors also expressed p53 should be noted, because WT1 has been shown to physically associate with and modify p53 action (35). Unlike p53, the expression of the c-ERB2 protooncogene was not correlated with either tumor grade or WT1 expression (data for clinical parameters is not shown).
The plus- and minus-KTS variants of WT1 have different DNA and RNA binding specificities and hence must transcriptionally control different constellations of genes (36, 37). In addition, a WT1 plus-KTS splice variant was recently shown to bind in a sequence-specific fashion to mRNA, implicating plus-KTS variants in post-transcriptional regulation of gene expression (28). Should WT1 be involved in gene-regulatory perturbations associated with breast cancer initiation or maintenance, we reasoned that tumor-related WT1 action on different sets of genes would be likely and could be deduced through detection of altered expression of KTS variants. For this reason we undertook a detailed analysis of the expression of the two classes of WT1 KTS variants (Figs. 2 and 3A) in tissue from the normal and cancerous breast.
Figure 2.
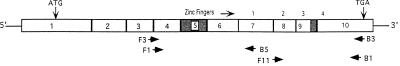
WT1 gene structure, splice variants, and detection scheme. The 10 exons of WT1 are shown as boxes and the alternatively spliced 51-base and 9-base sequences encoded by exon 5 and the 3′ end of exon 9 (KTS) are shaded. The location of the four zinc-finger motifs are noted above the boxes and the positions of the forward (F) and reverse (B) oligonucleotide primers relative to the transcript are shown.
Figure 3.
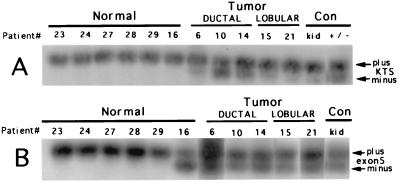
Detection and analysis of WT1 mRNA splice variants in normal (reduction mammoplasty) and cancerous breast tissue. (A) Reverse transcription-coupled PCR and Southern blot hybridization analysis of KTS splice variants for presence or absence of the 9-base KTS sequence. Upper row of signals, plus KTS; lower row, minus KTS. Normal (control) is kidney and a mixture of plus- or minus-KTS WT1-containing plasmids. Round 1 primers, F3/B3; round 2 primers, F11/B1. Two rounds of 30 and 25 amplification cycles for mammary gland WT1 mRNA; kidney WT1 mRNA could be detected with a single round of 30 cycles. (B) Reverse transcription-coupled PCR and Southern blot hybridization analysis of exon 5 splice variants. Upper row of signals, plus exon 5; lower row, minus exon 5. Normal (control) is kidney. Round 1 primers, F3/B3; round 2 primers, F1/B5.
By using nested pairs of PCR primers and Southern blot hybridization to detect low levels of target, a WT1-specific probe detected only the plus-KTS form(s) in samples from normal tissue from reduction mammoplasties, whereas, in addition to plus-KTS variants, minus-KTS signals of various intensities appeared for each tumor (Fig. 3A). As expected, both the plus- and minus-KTS forms were detected in the kidney and in the plasmid controls. Plus- and minus-exon-5 variants appeared as expected in all tumors and in the kidney control (Fig. 3B). In normal tissue, plus-exon-5 form(s) were the only ones expressed except for patient 16, where the minus form also appeared. Our splice-variant detection strategy was tested by coamplifying plasmids containing cDNA for either the WT1 message variant with both splices or neither splice. The appearance of amplification products of the predicted size validated the nested PCR system (Fig. 3A). Kidney tissue also expressed both splicing forms of KTS and exon 5 as predicted (Fig. 3) (21, 38).
A replicate PCR for the sample from patient 16 resulted in amplification of only the plus form, indicating that in some of our PCR trials, variants may not have been detected. In fact, when there exists a low abundance of two or more bone fide targets for a primer pair, as with the WT1 splice variants, a stochastic sampling error in which only a single target is amplified is likely if not inevitable. This pitfall has been studied in detail by Taberlet et al. (39), who demonstrated its avoidance by multiple PCR trials to detect all possible targets. The same study indicated that the frequency with which a target is detected will depend on its relative abundance; in multiple PCR trials, the more abundant targets will appear most frequently. This suggested to us that replicate amplifications could be used to quantify the relative proportions of the WT1 splice variants in tumor versus normal samples.
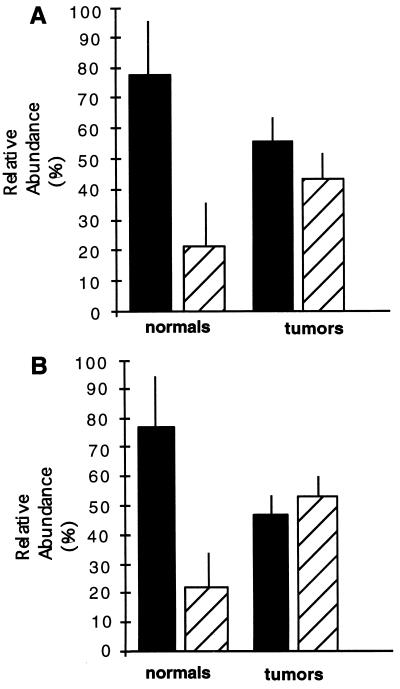
A multiple sampling scheme was, therefore, used to further investigate the proportions of KTS and exon 5 splice variants, the results of which were consistent with the initial PCR trial and add important details. Data on previously undetected splice variants showed that in normal breast tissue, the minus forms of each variant were present at significant levels, in the 20–30% abundance range (Fig. 4). In tumor tissue, this ratio was less pronounced due to a relative increase in the minus-KTS and minus-exon-5 variants to 40–50% of the splice variant mixture. Finally, breast tumors are often composed of a heterogeneous mixture of tumor and stromal cells. Although the latter must contribute to the WT1 mRNA pool, the relatively low levels of immunodetectable WT1 seen in the stroma and the random patient population, which should reflect a variety of epithelial/stromal ratios, suggest that systematic variation in stromal contribution between normal and tumor tissue is not likely to account for the observed differences in splice abundance.
Figure 4.
Reverse transcription-coupled PCR analysis of the relative proportion of KTS and exon 5 splice variants in normal versus cancerous breast tissue. (A) KTS variants. Bars: solid, plus forms; hatched, minus forms. (Total number of PCRs: normal, n = 31; tumor, n = 17). Normal tissue samples (n = 10) consisted of tissue from eight reduction mammoplasty patients and two samples of normal breast tissue in the vicinity of tumors. As a group and based on age (ranging from 23 to 45 years), these noncancer patients were considered premenopausal with the possible exception of one individual (52 years). Six tumor samples were analyzed. Data are the mean ± SD. (B) Exon 5 variants. Bars: solid, plus forms; hatched, minus forms. (Total number of PCRs: normal, n = 26; tumor, n = 15.)
DISCUSSION
The known tumor suppressor action of the WT1 gene in the developing kidney and its regulation of the genes encoding the mammotrophic IGF-IR/IGF-II system, as well as the mammary growth inhibitor TGF-β1, make it a candidate gene for action in the mammary gland. The goal of the current study was to determine whether WT1 was expressed in normal human breast tissue, and if so, whether this expression was altered in mammary tumors. Our key findings are that WT1 protein and mRNA are expressed in the normal breast and that levels and subcellular localization of WT1 protein and the alternative splicing of WT1 mRNA were significantly altered in a random sample of breast tumors (Figs. 1 and 3).
In the normal breast, the level and patterning of WT1 immunostaining strongly indicated that WT1 gene expression is differentially regulated between developmentally divergent ducts and lobules as well as within the mature duct. Duct lumenal cell staining contrasted with the lobular pattern, the former with a patchy distribution versus the latter with the more uniform distribution, suggesting that WT1 may play different developmental roles within these structures (Fig. 1 A and E). Developmental regulation of WT1 has been described in the mouse urogenital system and in rat ovarian development (29, 40).
Interestingly, the higher frequency of WT1-negative lobular tumors compared with ductal tumors suggests that developmental differences in expression may have sequelae in neoplasia. Within the duct, the distinctive differences in WT1 staining intensity between the myoepithelial and lumenal cell lineages and the subdivision of lumenal cells into WT1-positive and -negative populations encompassing putative stem- and differentiated-cell types are indicative of developmental regulation (Fig. 1A). The presence of WT1 protein in undifferentiated cells could be relevant both to normal development and tumorigenesis, as these cells are considered to be premitotic, poised to divide under the hormonal influences of pregnancy, for example. If we borrow from the Wilms tumor model, where ablation of WT1 action results in derepression of the IGF-IR and IGF-II (25), then in the mammary gland, suppression of this mammotrophic autocrine–paracrine loop by WT1 would normally maintain these cells in mitotic arrest and loss or inactivation of WT1 would lead to unregulated cell division and tumors.
The most striking features of WT1 immunostaining in breast tumors were (i) the absence of immunodetectable WT1 in a majority of tumor cells (Fig. 1 C and F and Table 1), (ii) cytoplasmic localization in a subset of tumor cells (Fig. 1 B and D), and (iii) high levels in some advanced tumors, suggesting possible overexpression (Fig. 1D and Table 1). All are indicative of breast-tumor-related perturbations of WT1 expression. Since significant reduction of WT1 mRNA was not detected in breast tumors (Fig. 3), the tumor-related changes in expression patterns of the alternatively spliced WT1 mRNA species must in part underlie the changes in protein expression noted above (Figs. 3 and 4). Consistent with this idea, in all tumors studied, the normal proportions of the splice variants were replaced with increased proportions of both minus-exon-5 and minus-KTS variants (Fig. 4). The appearance of this pattern in a random sample of breast tumors suggests that altered splicing of WT1 mRNA is characteristic of breast tumor tissue, potentially resulting in a change in the set of target genes subject to WT1 regulation (Fig. 3). The fact that the observed pattern was shared by ductal and lobular tumors derived from developmentally divergent tissues may mean that a cell type common to both tissues (e.g., an early progenitor cell) was affected prior to differentiation (Fig. 3).
General loss of WT1 protein was evident in a cancerous versus normal lobular tissue sample from the same patient (Fig. 1 E and F, respectively). The origin of WT1-negative ductal tumor cells is a more complex issue, given that normal ductal epithelium contains many WT1-negative cells. If the latter were selectively vulnerable to neoplastic transformation, however, this would still be consistent with a tumor suppressor function for WT1 in the normal breast.
In either case, whether nuclear WT1 expression is actively lost or WT1-negative cells are selected, the establishment of the WT1-negative tumor phenotype can occur early in tumorigenesis. Thus, in a carcinoma in situ, the incipient tumor was negative for nuclear WT1 while still contained in apparently normal duct (Fig. 1B), indicating that the WT1-negative status of the derivative frank carcinoma (Fig. 1C) was established at its inception. The persistence of the WT1-negative phenotype in this case would be consistent with continued tumor growth requiring the reduction or absence of WT1 expression.
The discovery of cytoplasmic, but not nuclear, WT1 in the cells of three tumors (Fig. 1 B, D, and F) suggests that inactivation of WT1 may occur by restricting its access to nuclear targets. Recent experiments have shown that cytoplasmic retention inhibits the normal regulatory functions of WT1 (27). Phosphorylation is a major post-translational mechanism regulating transcription factor action, and phosphorylation of WT1 by protein kinase A resulted in the cytoplasmic retention of WT1 protein, inhibiting the transcriptional suppressor activity of a WT1 reporter construct in 3T3 cells. Interestingly, in mammary epithelial cells in vitro, protein kinase A activation by cholera toxin stimulated proliferation (41) and, in vivo, locally elevated cAMP levels powerfully stimulated the growth of regressed ducts in the mouse mammary gland (42), effects that are consistent with the inhibition of a growth suppressor. Cytoplasmic retention has recently been suggested as a mechanism for the regulation of other transcription factors. The breast tumor suppressor gene product BRCA1, a nuclear phosphoprotein and putative transcription factor, is aberrantly localized to the cytoplasm in most breast cancer cells (27, 43), and in oligodendrocytes in vitro, the p53 tumor suppressor protein moves to the cytoplasm during proliferation and shifts to the nucleus during differentiation where it mediates differentiation or apoptosis (44). If we return to our observations on the subcellular localization of WT1, we conclude that mammary cells with cytoplasmic WT1 are likely to be under a different program of gene regulation than their counterparts with nuclear protein localization.
Finally, the higher-grade most-dangerous tumors showed interesting correlations among WT1 expression, estrogen receptor status, and p53 expression. In estrogen-receptor-positive tumors, the majority of cells lacked WT1 in 78% of the cases; however, in the more-advanced estrogen-receptor-negative counterparts, the majority of cells lacked WT1 in only 40% of the cases (data not shown). In the latter, higher average staining levels suggested possible negative regulation of tumor WT1 by estrogen. In addition, there was a correlation in higher-grade tumors between WT1 and p53 expression. Recent cotransfection studies have shown that WT1 can stabilize p53 protein and inhibit p53-mediated apoptosis (35). In breast tumors, overexpression of p53, usually associated with its functional inactivation, is correlated with high levels of IGF-IR, whereas normal p53 protein repressed IGF-IR expression (45, 46). Overexpression of WT1 or, possibly, perturbation of WT1 splice variant expression in breast tumors might, therefore, interfere with the normal surveillance activities of p53. Conversely, p53 might under certain circumstances, interfere with normal WT1 action(s). Expression of c-ERB2, an epidermal growth factor receptor-related oncogene, was not obviously correlated with either tumor status or WT1.
In conclusion, a role for WT1 in mammary development is inferred from its differential localization in lobules versus ducts, as well as within ducts, where it was present in putative progenitor cells. With regard to WT1 and breast tumorigenesis, if WT1 regulates the IGF and TGF-β systems in mammary epithelium, then our observations indicate that it would be a candidate for a breast cancer tumor suppressor gene. This conclusion is based on the following points: (i) a high percentage of breast cancer cells lacked immunodetectable WT1, indicating that WT1 could be responsible for the overexpression of IGF-IR common to breast cancers; (ii) cytoplasmic localization of WT1 suggests that functional inactivation of WT1 occurs in a subset of tumors; (iii) nuclear WT1 protein was absent in tumors in situ, suggesting that altered WT-1 expression may coincide with the first appearance of tumor cells possibly reflecting an event related to cause; (iv) the WT1-negative phenotype was maintained in tumor masses, consistent with a requirement for on-going lack of expression in growing tumors; (v) common tumor-related perturbations of mRNA splice usage were demonstrated.
Given our data, it should now be of interest to elucidate those mechanisms that determine WT1 function in the normal and cancerous human breast and the possible role of WT1-regulated IGF and TGF-β action in mammary growth and tumorigenesis.
Acknowledgments
We thank J. Snyder, M.D. and Kelly R. O’Keefe, M.D., Ph.D. (Dominican Hospital, Santa Cruz); Mr. D. Albritton, Ms. P. Huerta, and Ms. E. Olson (University of California, Santa Cruz); and Dr. Stephen Ethier (University of Michigan Breast Tissue Bank) for obtaining breast tissue samples. C.T.R. thanks Karen Pacelli for expert technical assistance. This work was supported by National Institutes of Health Grants DK-48883 and HD-27845 (to C.W.D.) and DK-50810 (to C.T.R.).
ABBREVIATIONS
- IGF
insulin-like growth factor
- IGF-IR
IGF-I receptor
- TGF-β
transforming growth factor β
References
- 1.Ruan W, Catanese V, Wieczorek R, Feldman M, Kleinberg D. Endocrinology. 1995;136:1296–1302. doi: 10.1210/endo.136.3.7867584. [DOI] [PubMed] [Google Scholar]
- 2.Yee D. Breast Cancer Res Treat. 1994;32:85–95. doi: 10.1007/BF00666209. [DOI] [PubMed] [Google Scholar]
- 3.Lee A, Yee D. Biomed Pharmacother. 1995;49:415–421. doi: 10.1016/0753-3322(96)82678-3. [DOI] [PubMed] [Google Scholar]
- 4.Peyrat J, Bonneterre J. Breast Cancer Res Treat. 1992;22:59–67. doi: 10.1007/BF01833334. [DOI] [PubMed] [Google Scholar]
- 5.Brunner N, Yee D, Kern F, Spang-Thomsen M, Lippman M, Cullen K. Eur J Cancer. 1993;29A:562–569. doi: 10.1016/s0959-8049(05)80152-2. [DOI] [PubMed] [Google Scholar]
- 6.Cullen K J, Yee D, Sly W S, Perdue J, Hampton B, Lippman M E, Rosen N. Cancer Res. 1990;50:48–53. [PubMed] [Google Scholar]
- 7.Kleinberg D. J Mammary Gland Biol Neoplasia. 1997;2:49–58. doi: 10.1023/a:1026373513521. [DOI] [PubMed] [Google Scholar]
- 8.Silberstein G B, Daniel C W. Science. 1987;237:291–293. doi: 10.1126/science.3474783. [DOI] [PubMed] [Google Scholar]
- 9.Daniel C W, Silberstein G B, Van Horn K, Strickland P, Robinson S. Dev Biol. 1989;135:20–30. doi: 10.1016/0012-1606(89)90154-1. [DOI] [PubMed] [Google Scholar]
- 10.Silberstein G B, Flanders K C, Roberts A B, Daniel C W. Dev Biol. 1992;152:354–362. doi: 10.1016/0012-1606(92)90142-4. [DOI] [PubMed] [Google Scholar]
- 11.MacCallum J, Bartlett J, Thompson A, Keen J, Dixon J, Miller W. Br J Cancer. 1994;69:1006–1009. doi: 10.1038/bjc.1994.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker R, Dearing S, Gallacher B. Br J Cancer. 1994;69:1160–1165. doi: 10.1038/bjc.1994.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arteaga, C., Dugger, T., Winnier, A. & Forbes, J. (1993) J. Cell. Biochem. 17G, Suppl., 187–193. [DOI] [PubMed]
- 14.Arteaga C, Hurd S, Winnier A, Johnson M, Fendly B, Forbes J. Clin Invest. 1993;92:2569–2576. doi: 10.1172/JCI116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond I, Madden S, Rohwer-Nutter P, Bell G, Sukhatme V, Rauscher F J., III Science. 1992;257:674–677. doi: 10.1126/science.1323141. [DOI] [PubMed] [Google Scholar]
- 16.Werner H, Re G G, Drummond I A, Sukhatme V P, Rauscher F J, III, Sens D A, S, Garvin A J, LeRoith D, Roberts C T., Jr Proc Natl Acad Sci USA. 1993;90:5828–5832. doi: 10.1073/pnas.90.12.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dey B, Sukhatme V P, Roberts A B, Sporn M B, Rauscher F J, III, Kim S-J. Mol Endocrinol. 1994;8:595–602. doi: 10.1210/mend.8.5.8058069. [DOI] [PubMed] [Google Scholar]
- 18.Werner H, Rauscher F J, III, Sukhatme V, Drummond I, Roberts C T, Jr, LeRoith D. J Biol Chem. 1994;269:12577–12585. [PubMed] [Google Scholar]
- 19.Haber D, Buckler A. New Biol. 1992;4:97–106. [PubMed] [Google Scholar]
- 20.Rauscher F J., III FASEB J. 1993;7:896–903. [PubMed] [Google Scholar]
- 21.Haber D, Sohn R, Buckler A, Pelletier J, Call K, Housman D. Proc Natl Acad Sci USA. 1991;88:9618–9622. doi: 10.1073/pnas.88.21.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haber D, Housman D. Adv Cancer Res. 1992;59:41. doi: 10.1016/s0065-230x(08)60302-4. [DOI] [PubMed] [Google Scholar]
- 23.Coppes M, Campbell C, Williams B. FASEB J. 1993;7:886–895. doi: 10.1096/fasebj.7.10.8393819. [DOI] [PubMed] [Google Scholar]
- 24.Huff V, Saunders G. Biochim Biophys Acta. 1993;1155:295–306. doi: 10.1016/0304-419x(93)90011-z. [DOI] [PubMed] [Google Scholar]
- 25.Roberts C T., Jr Ann Intern Med. 1995;122:57–58. doi: 10.7326/0003-4819-122-1-199501010-00009. [DOI] [PubMed] [Google Scholar]
- 26.Larsson S, Charlieu J, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie N. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- 27.Ye Y, Raychaudhuri B, Gurney A, Campbell C, Williams B. EMBO J. 1996;15:5606–5615. [PMC free article] [PubMed] [Google Scholar]
- 28.Caricasole A, Duarte A, Larsson S H, Hastie N D, Little M, Holmes G, Todorov I, Ward A. Proc Natl Acad Sci USA. 1996;93:7562–7566. doi: 10.1073/pnas.93.15.7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu S, Kubo M, Sang-Young C, Haluska F, Housman D, Hsueh A. Mol Endocrinol. 1995;9:1356–1366. doi: 10.1210/mend.9.10.8544844. [DOI] [PubMed] [Google Scholar]
- 30.Smith G H, Medina D. J Cell Sci. 1988;89:173–183. doi: 10.1242/jcs.90.1.173. [DOI] [PubMed] [Google Scholar]
- 31.Smith G H. Breast Cancer Res Treat. 1996;39:29–31. doi: 10.1007/BF01806075. [DOI] [PubMed] [Google Scholar]
- 32.Williams J M, Daniel C W. Dev Biol. 1983;97:274–290. doi: 10.1016/0012-1606(83)90086-6. [DOI] [PubMed] [Google Scholar]
- 33.Silberstein G B, Van Horn K, Shyamala G, Daniel C. Cell Growth Differ. 1996;7:945–952. [PubMed] [Google Scholar]
- 34.Barnes D, Camplejohn R. J Mammary Gland Biol Neoplasia. 1996;1:163–175. doi: 10.1007/BF02013640. [DOI] [PubMed] [Google Scholar]
- 35.Maheswaran S, Englert C, Bennett P, Heinrich G, Haber D. Genes Dev. 1995;9:2143–2156. doi: 10.1101/gad.9.17.2143. [DOI] [PubMed] [Google Scholar]
- 36.Rauscher F J, III, Morris J, Tournay O, Cook D, Durran T. Science. 1990;250:1259–1262. doi: 10.1126/science.2244209. [DOI] [PubMed] [Google Scholar]
- 37.Drummond I, Rupprecht H, Rohwer-Nutter P, Lopez-Guisa J, Madden S, Rauscher F J, III, Sukhatme V. Mol Cell Biol. 1994;14:3800–3809. doi: 10.1128/mcb.14.6.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenner B, Wildhardt G, Schneider S, Royer-Pokora B. Oncogene. 1992;7:1431–1433. [PubMed] [Google Scholar]
- 39.Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits L, Bouvet J. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelletier J, Schalling M, Buckler A, Rogers A, Haber D, Housman D. Genes Dev. 1991;5:1345–1356. doi: 10.1101/gad.5.8.1345. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Guzman R, Richards J, Imagawa W, McCormick K, Nandi S. Endocrinology. 1980;107:35–41. doi: 10.1210/endo-107-1-35. [DOI] [PubMed] [Google Scholar]
- 42.Silberstein G B, Strickland P, Trumpbour V, Coleman S, Daniel C W. Proc Natl Acad Sci USA. 1984;81:4950–4954. doi: 10.1073/pnas.81.15.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C, Li S, Chen Y, Chen P, Sharp Z, Lee W. J Biol Chem. 1996;271:32863–32868. doi: 10.1074/jbc.271.51.32863. [DOI] [PubMed] [Google Scholar]
- 44.Eizenberg O, Faber-Elman F, Gottlieb E, Oren M, Rotter V, Schwartz M. Mol Cell Biol. 1996;16:5178–5185. doi: 10.1128/mcb.16.9.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werner H, Karnieli E, Rauscher F J, III, LeRoith D. Proc Natl Acad Sci USA. 1996;93:8318–8323. doi: 10.1073/pnas.93.16.8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster N, Resnik J, Reichart D, Strauss B, Haas M, Seely L B. Cancer Res. 1996;56:2781–2788. [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. 6.7. [Google Scholar]