Abstract
Many signaling and trafficking proteins contain modular domains that bind reversibly to cellular membranes. The structural basis of the intermolecular interactions which mediate these membrane-targeting events remains elusive since protein-membrane complexes are not directly accessible to standard structural biology techniques. Here we report a fast protein-micelle docking methodology that yields three-dimensional model structures of proteins inserted into micelles, revealing energetically favorable orientations, convergent insertion angles, and an array of protein-lipid interactions at atomic resolution. The method is applied to two peripheral membrane proteins, the early endosome antigen 1 (EEA1) FYVE (a zinc finger domain found in the proteins Fab1, YOTB/ZK632.12, Vac1, and EEA1) and Vam7p phagocyte oxidase homology domains, which are revealed to form extensive networks of interactions with multiple phospholipid headgroups and acyl chains. The resulting structural models explain extensive published mutagenesis data and reveal novel binding determinants. The docking restraints used here were based on NMR data, but can be derived from any technique that detects insertion of protein residues into a membrane, and can be applied to virtually any peripheral membrane protein or membrane-like structure.
INTRODUCTION
A large number of soluble proteins are transiently recruited to biological membranes during various cellular processes, including cell signaling and membrane trafficking. They form a special class, commonly referred to as peripheral membrane proteins, and use different mechanisms for reversible membrane attachment (1). Some of these proteins have modular domains that associate specifically with the headgroups of their membrane-embedded lipid ligands, such as phagocyte oxidase homology (PX) and FYVE (a zinc finger domain found in the proteins Fab1, YOTB/ZK632.12, Vac1, and early endosome antigen 1) domains, which are specialized for phosphoinositide binding. Other peripheral membrane proteins have covalently attached lipid anchors, such as Ras proteins, which are recruited to membranes through acylated C-terminal CAAX (C, Cys; A, an aliphatic amino acid; X, any amino acid) box motifs. Alternatively, some enzymes utilize part of their molecular surface for bilayer interactions, such as phosphatidylinositol-specific phospholipase C, which is attached to membranes by a cluster of hydrophobic residues located close to its catalytic site. In addition, many receptor-soluble domains, such as regulatory protein subunits of ion channels and transmembrane receptors, are thought to bind weakly or dynamically to membranes.
Several experimental methods have been used to characterize the membrane interactions of peripheral proteins, including electronic paramagnetic resonance spectroscopy (2–5), site-directed fluorescence (4,6,7), monolayer penetration (8–10), x-ray reflectivity studies (11,12), and solid state (13–16) and solution NMR spectroscopy (17–20). These techniques provide complementary quantitative and qualitative insights into the mechanisms of membrane interaction, membrane docking geometry, and the composition of the binding sites. However they rarely resolve the fine details of intermolecular interface with specific lipid ligands and the surrounding membrane surface. Recent molecular dynamics (MD) studies attempt to fill this gap by providing an all-atom picture of membrane binding domains embedded in phospholipids bilayers (21,22). Besides the large computational cost, a major drawback of this approach lies in the difficulty of incorporating the available experimental data to accurately represent the respective molecular orientations.
A simple way of probing protein-membrane interactions by solution NMR spectroscopy is the detection of chemical shift changes of a protein's amide signals upon titration of a micelle in a series of [1H,15N] heteronuclear single quantum coherence (HSQC) experiments (23). The main limitation of using chemical shifts is the possibility of indirect effects which can obscure the detection of protein-micelle interfaces. Other NMR experiments can be utilized to detect proximities between protein and micelle nuclei. Under favorable exchange conditions, nuclear Overhauser effect (NOE) spectroscopy experiments can be employed to obtain short-range intermolecular NOE distances (24). For longer range interactions, paramagnetic relaxation enhancement (PRE) of protein resonances caused by a spin-labeled micelle can be measured (25). In addition, transferred cross-saturation experiments can be used to investigate the relaxation transfer across the protein-bilayer interface (26).
Here we present a novel method which makes use of NMR data obtained from protein-micelle complexes (or any other type of experimental evidence of membrane interaction sites) to drive the docking of a protein structure to an MD model of a micelle while accounting for the inherent flexibility of the interfaces. This method is applied to two structurally distinct peripheral membrane proteins, the FYVE domain of early endosome antigen 1 (EEA1) and the PX domain of the Vam7p target soluble N-ethylmaleimide sensitive factor attachment protein receptor, for which protein-micelle NMR interaction data are available (27,28). The experimentally based docking models reveal for the first time, to our knowledge, not only the array of recognition determinants for phospholipids and membrane surfaces but also the three-dimensional structures of the two peripheral membrane proteins inserted into membrane-like micelles.
METHODS
NMR data treatment
Here we primarily relied on PREs to detect the positions of residues inserted into micelles. A recent publication suggested a quantitative treatment of intermolecular PRE of NMR signals of residues inserted within doxyl-labeled micelles by assuming a radial Gaussian distribution of the spin label inside an idealized spherical micelle (29). Under the further assumptions of slow intra- and intermolecular dynamics as compared with the correlation time of the complex and for a rigid protein structure, the Solomon equation (30) could be integrated, which in turn allowed the estimation of the protein's insertion angle in the micelle. In principle, the minimization procedure should also allow the estimation of distances between the corresponding protein protons and the micelle center. However, this approach did not lead to reliable intermolecular distances, possibly due to a non-Gaussian distribution of the spin label inside the micelles (Supplementary Fig. S1) and sizable effects caused by internal dynamics and spin label flexibility. Another method employs a model-free approach in the Solomon theory to describe the local dynamics of flexible paramagnetic centers (31), which, for this particular system, is hampered by the large conformational space sampled by the lipid-attached spin label in the micelle.
We have developed a new method which offers improved structural accuracy, computational efficiency, and general applicability by incorporating different experimental data types into semiquantitative restraints between individual protein residues and the micelle center. The distance restraint boundaries are estimated from the radial distribution of the micelle atoms based on an MD model for the micelle (Supplementary Fig. S2). Protein residues are considered to insert into the micelle interior if i), buried spin labels (e.g., 14-doxyl) cause paramagnetic enhancements in their resonances, ii), they exhibit intermolecular NOEs to the detergent acyl chains, and iii), their chemical shifts are perturbed upon micelle addition. These residues are restrained inside the micelle to any position that is below the area of the micelle phosphate groups. Similarly, protein residues interacting with the micelle surface are defined as those with i), resonances which are affected by an interfacial spin label (e.g., 5-doxyl), ii), chemical shifts which are perturbed by micelle association, and iii), a lack of observable intermolecular NOEs to micelle acyl chains. The positions of the latter class of residues were restrained to below the region occupied by the terminal headgroups, in this case the choline group of the dodecylphosphocholine (DPC) molecule.
Semiquantitative restraints can be used to drive docking of protein-micelle complexes by an extension of the HADDOCK program (32) in a fashion similar to protein-protein docking. The protein-micelle docking approach requires an experimental or homology-modeled protein structure and an MD model of the micelle. It then invokes protein-micelle interaction restraints to search for energetically favorable intermolecular contacts by simulated annealing and restrained MD. Simulations are performed on a large number of starting points consisting of a spatially randomized ensemble of micelle and protein structures to sample a large micelle surface and to account for any conformational variability of the complex. Flexibility is allowed only for the protein residues likely to interact with the micelle based on the experimental evidence, and it is gradually introduced, first for side chains and then for both side-chain and backbone atoms in the protein-micelle interface. The large number of parallel simulations requires fast computations, which are achieved by vacuum torsion angle dynamics (33). The best models are then subjected to Cartesian MD in explicit solvent, which provide realistic nonbonded interactions for accurate energy-based rankings of the docking models.
Protein and micelle structures
The structures of the EEA1 FYVE domain bound to inositol 1,3-bisphosphate [Ins(1,3)P2] and the uncomplexed Vam7p PX domain were obtained from Protein Data Bank files 1JOC (34) and 1KMD (35), respectively. For the FYVE domain, only the monomer unit was considered, as the protein is largely monomeric in the presence of DPC micelles (27). The starting structure of the DPC micelle was constructed based on an equilibrated MD model of the DPC micelle containing 54 detergent molecules (36), which closely matches the aggregate number in the FYVE-DPC complex estimated by pulsed field gradient NMR experiments (27). Topology and parameter files of the DPC molecule for the CNS program (37) were generated with PRODRG (38) and manually adjusted to include nonpolar hydrogen atoms and to match the PARALLHG 5.3 force field (39) and optimized potential for liquid simulations (OPLS) nonbonded parameters (40). Partial charge distribution was taken from the MD study. The model was adapted to the new force field and nonbonded parameters by immersing the micelle into an 8 Å shell of TIP3P water molecules (41) followed by 200 steps of steepest descent energy minimization, 500 MD heating steps at 100 and 200 K, and 5500 MD steps at 300 K, all with a 2 fs time step. This initial equilibration continued with a productive simulation when 20 models were sampled after every 5000 steps of the MD trajectory.
The structure of the Vam7p PX domain complexed with Ins(1,3)P2 was obtained by restrained ligand docking with the program HADDOCK (version 1.3). Two absolutely conserved basic residues, Arg-41 and Arg-88, form hydrogen bonds with the PtdIns(3)P headgroup based on structural homology with the PtdIns(3)P-bound PX domains of p40phox (42) and Grd19p (43) and NMR chemical shift changes induced by ligand titration (28). Ambiguous hydrogen bonds of 1.8–2.3 Å (hydrogen acceptor) and 2.7–3.3 Å (donor acceptor) between the side-chain donors of the two conserved arginine residues and the predicted acceptors of the Ins(1,3)P2 ligand were used as distance restraints during docking. All-atoms topology and parameter files for the Ins(1,3)P2 ligand together with an energy minimized structure were obtained with XPLO2D (Uppsala Software Factory, Uppsala, Sweden) and PRODRG. The 20 published NMR structures of the free-state Vam7p PX domain were used to generate 800 random orientations of the protein-ligand complex, which were subsequently refined by restrained rigid body docking. The best 400 models with respect to the intermolecular energies (the sum of electrostatic, van der Waals, and ambiguous hydrogen-bond restraints) were passed to the second stage of semiflexible refinement where the ligand was left fully flexible and the protein flexibility was gradually introduced along side chains and backbone at the protein-ligand interface. The flexible interface spanned Tyr-26–Tyr-29, Tyr-39–Arg-51, Asp-63–Asp-81, and Met-84–Glu-93 based on induced chemical shift variations in these elements. In the final stage, the 200 lowest energy models were refined in explicit solvent (i.e., water), and the best 10 models were chosen as starting structures for protein-micelle docking.
Docking protocol
Protein-micelle interaction restraints used for docking were defined as distance restraints between the backbone amide protons of the protein and the geometric center of the micelle employing a center averaging over the micelle heavy atoms and a soft-square restraining potential in CNS. NMR experimental data were used to distinguish two classes of residues: deeply inserting residues (that is, below the phosphate groups) and interfacially active residues (that is, below the choline groups). The backbone amide protons of deeply inserted and interfacial residues were restrained to within 20 Å and 22 Å from the micelle center, respectively. These distances were based on the radial distributions of the phosphate and choline groups in the MD model structures of the DPC micelle. Specifically, the DPC phosphorous and nitrogen atoms occupy positions that are typically below 20 Å and 22 Å from the micelle center (i.e., their mean positions plus one standard deviation, see Supplementary Fig. S2). Protein-micelle docking was achieved by an extension of the standard protein-protein docking protocol with HADDOCK. The docking protocol comprises the three stages: i), complex generation and orientational optimization, ii), semiflexible docking, and iii), refinement in explicit solvent. The primary differences from the standard HADDOCK method are the development of protein-micelle restraints and linked DPC molecules to allow fast torsion angle dynamics of micelles.
Complex generation and optimization
Twenty MD structures of the DPC micelle together with the initial protein structures (1 crystal structure for FYVE or the 10 NMR-derived structures for the PX domain) were used to create 400 combinations of randomly positioned protein-micelle complexes separated by 5 Å. Each complex was optimized by rotational rigid body minimization to ensure the proper orientation of the molecules before docking. Only the protein was allowed to rotate; each minimization attempt was repeated five times, and the target function was based solely on the distance restraint's term.
Semiflexible docking
The 400 structures of each protein-micelle complex were subjected to a semiflexible docking stage comprising three phases of MD-simulated annealing in torsion angle space: i), rigid body simulated annealing (3000 MD steps at 2000 K and 2000 MD cooling steps to 500 K), ii), semiflexible simulated annealing (2000 steps from 1000 to 50 K) where the side chains of the protein residues at the interface and the complete DPC micelle were allowed to move, and iii), semiflexible simulated annealing part (2000 MD steps from 500 to 50 K) where side chains and the backbone of the protein residues at the interface and the complete micelle molecule were allowed to move. To enable fast torsion angle dynamics for the micelle, the DPC molecules were artificially linked by strings of five dummy atoms with no steric restrictions imposed on the micellar aggregate (that is, zero van der Waals radii, no partial charges, and bond lengths of 5 Å within the string and 7 Å to the methyl carbons of sequential acyl chains). In a similar fashion the Ins(1,3)P2 ligand was connected to the protein by a 3 Å artificial bond replacing a donor-acceptor hydrogen bond. Additionally, in the case of the FYVE domain, the two zinc ions were each connected by a 2.3 Å bond to the sulfur atom of a neighboring cysteine residue.
Refinement in explicit solvent
The final refinement is based on Cartesian dynamics performed in a thin shell of TIP3P water molecules. Position restraints were imposed on the protein heavy atoms outside the flexible interface, whereas all micelle atoms were left fully flexible. As allowed by the Cartesian dynamics, the artificial linkages of the DPC molecules, Ins(1,3)P2 ligand, and Zn+2 ions were discarded.
Models analysis
The docking solutions were ranked based on the sum of intermolecular electrostatic, van der Waals, and distance restraints energies. The nonbonded energies were evaluated using full electrostatic and van der Waals energy terms with an 8.5 Å distance cutoff, OPLS nonbonded parameters, and a dielectric constant of 10 during the in vacuo part of the protocol. The intermolecular hydrogen bonds between the protein and the phosphate groups of the DPC micelle and Ins(1,3)P2 ligand were evaluated with the program MOLMOL (44) using a hydrogen-acceptor distance cutoff of 2.5 Å and a maximum hydrogen-donor-acceptor angle of 35°. Salt bridges between Asp and Glu carboxylates and choline amine groups were considered to be significant if the distance between the corresponding oxygen and nitrogen atoms was smaller than 3.9 Å. The protein-micelle hydrophobic contacts were considered for methyl/methylene carbons distances shorter than 3.9 Å. The micelle center coordinates in the inertia tensor frame of the protein were calculated in MATLAB (The MathWorks, Natick, MA). Solvent accessibilities of protein atoms were estimated with Naccess (45). Molecular representations were generated with MOLMOL.
RESULTS AND DISCUSSION
FYVE-DPC complex
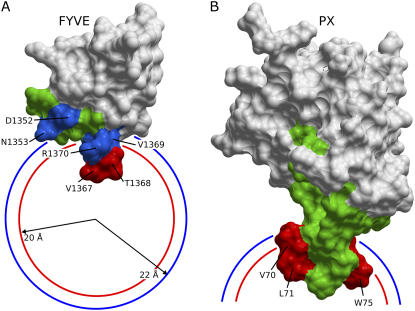
Various NMR experiments were previously used to collect data on the interactions of the FYVE domain with micelles, including chemical shift perturbations, intermolecular NOEs, and PREs induced by micelle-incorporated spin labels (Table 1). Two residues, Val-1367 and Thr-1368, insert into the micelle as they exhibit intermolecular NOEs to the methylene protons of the DPC acyl chain, strong backbone amide PREs caused by a phosphatidylcholine derivative carrying a 14-doxyl spin label near the micelle center and large DPC-induced chemical shift changes (27). The PREs caused by the interfacially active 5-doxyl spin label and the chemical shift changes induced by DPC indicate that Asp-1352, Asn-1353, Val-1355, Phe-1365, Ser-1366, Val-1369, Arg-1370, and Cys-1382 interact with the micelle surface (18,27). Hence, altogether there are two deep and eight shallow residues within the protein-micelle interface which contribute intermolecular restraints (Fig. 1 A).
TABLE 1.
Experimental data used for the definition of protein-micelle interaction restraints
| System | Experimental data | Residues | Reference |
|---|---|---|---|
| FYVE-DPC | Chemical shift perturbations | A1350, D1352, N1353, V1355, Q1356, F1365, S1366, V1367, T1368, V1369, R1370, C1382, A1383 | (18) |
| Intermolecular NOEs | V1367, T1368 | (27) | |
| 5-doxyl intermolecular PREs | D1352, N1353, V1355, F1365, S1366, V1367, T1368, V1369, R1370, R1371, C1382 | (27) | |
| 14-doxyl intermolecular PREs | V1367, T1368, V1369, R1370, H1373, I1380, F1406 | (27) | |
| PX-DPC | Chemical shift perturbations | E66, V70, L71, R73, R74, W75, R77, Y79 | This study |
| PX-DHPC | Chemical shift perturbations | E66, K67, V70, L71, R73, R74, W75, Q76, R77 | (28) |
| 10-doxyl PREs | V70, L71, W75 | (28) |
FIGURE 1.
Interaction restraints used for docking the FYVE-DPC and PX-DPC complexes. A and B show the solvent accessible areas of the FYVE and PX structures. Deeply inserting residues (below phosphate groups) are colored in red, interfacial interacting residues (below the micelle surface) are colored in blue, and the additional residues left flexible during docking are colored in green. Visible residues are indicated by arrows. The average positions of the phosphate and choline groups in the DPC micelle (20 and 22 Å from the micelle center) are depicted as red and blue circle sections, respectively.
The interaction restraints were used to dock the FYVE-DPC complex starting from the x-ray crystal structure of the FYVE domain bound to Ins(1,3)P2 and 20 MD model structures of the DPC micelle (see Methods section). The intermolecular hydrogen bonds between the Ins(1,3)P2 ligand and residues Asp-1352, Arg-1370, His-1372, His-1373, and Arg-1375 seen in the crystal structure were maintained during docking by imposing 1.8–2.3 Å and 2.8–3.3 Å distance restraints between the hydrogen-acceptor and the donor-acceptor atoms. Similarly, the tetrahedral coordination of the two zinc ions was preserved by imposing upper limits of 2.3 Å to the sulfur atoms of the four neighboring cysteine residues. Flexibility within the protein-micelle interface during docking was restricted to Glu-1351–Gln-1356 and Gly-1364–Arg-1371 based on their chemical shift sensitivities to DPC and their solvent accessibilities.
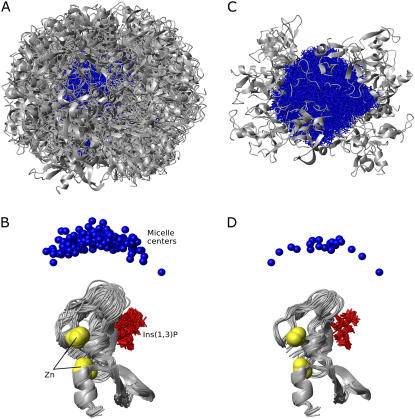
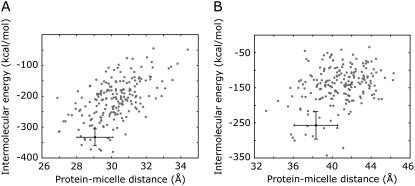
The protein populates essentially the entire micelle surface, as seen from the 200 models that were generated upon water refinement (Fig. 2 A). Due to the approximately spherical symmetry of the micelle, the relative orientation of the complex units can be defined by the position of the micelle center with respect to the protein (Fig. 2 B). The structural statistics of the energetically best 20 models (Figs. 2, C and D, and 3 A) are presented in Table 2. The ensemble is well defined with an average nonbonded intermolecular energy of the best 20 structures of 339.0 ± 43.7 kcal/mol, an average backbone root mean-square deviation (RMSD) of the protein interface of 0.62 ± 0.22 Å, and an average deviation of the micelle centers from the mean position after superimposing the protein backbones of 6.3 ± 4.2 Å. The quality of the docking models and the completeness of the interaction data set were assessed by twofold cross-validation. Similar RMSD values for excluded restraints and nonbonded intermolecular energies were found for the cross-validation and original runs, confirming the validity of the method for docking protein-micelle models using a small number of interaction restraints (see Supplementary Table S3).
FIGURE 2.
Selection of the FYVE-DPC complex structures. The upper and lower panels show the protein-micelle complexes after superimposing the micelle heavy atoms and protein backbones, respectively. A and B depict the 200 water refined structures, and C and D show the energetically best 20 structures. The proteins are represented as gray ribbons, micelles as blue sticks, micelle centers as blue balls, zinc ions as yellow spheres, and Ins(1,3)P2 as red sticks.
FIGURE 3.
Intermolecular energies of the FYVE-DPC and PX-DPC complexes. Plot of the intermolecular energies as a function of the distances between protein and micelle centers for (A) FYVE-DPC and (B) PX-DPC complexes. The solid circle and bars represent the average values and the corresponding standard deviations of the energetically best 20 structures.
TABLE 2.
Structural statistics of the best 20 model structures of FYVE-DPC/PX-DPC complexes
| RMSD from the mean protein structure (Å) | |
| Flexible interface backbone* | 0.62 ± 0.22/1.34 ± 0.27 |
| All backbone | 0.75 ± 0.21/1.56 ± 0.29 |
| RMSD from the mean protein initial structure (Å)† | |
| Flexible interface backbone | 0.99 ± 0.29/1.86 ± 0.32 |
| All backbone | 1.08 ± 0.32/1.82 ± 0.44 |
| Number of interaction restraints | 10/3 |
| Number of flexible residues | 14/23 |
| Intermolecular energies after water refinement (kcal/mol) | |
| Evdw | −79.3 ± 11.8/−63.2 ± 10.4 |
| Eelec | −259.7 ± 31.9/−194.0 ± 37.0 |
| Erestraints | 6.2 ± 2.3/0.3 ± 0.8 |
| Buried surface area (Å2) | 1622.7 ± 166.2/1279.3 ± 197.8 |
| RMSD from idealized covalent geometry | |
| Bonds (Å) | 0.003 ± 0.000/0.003 ± 0.000 |
| Angles (deg) | 1.658 ± 0.002/0.972 ± 0.156 |
| Impropers (deg) | 1.402 ± 0.007/1.178 ± 0.005 |
| Ramachandran plot (%) | |
| Allowed regions | 89.3/100.0 |
| Disallowed regions | 0.7/0.0 |
| Micelle center coordinates in the protein inertia frame‡ | |
| θ (deg) | 24.7 ± 11.4/38.7 ± 6.4 |
| ψ (deg) | 196.1 ± 60.2/77.6 ± 12.7 |
| r (Å) | 29.0 ± 1.0/38.3 ± 2.2 |
| Deviation of micelle centers from the mean (Å)§ | 6.3 ± 4.2/6.5 ± 3.6 |
| RMSD from the mean micelle structure (Å) | 2.84 ± 0.29/2.96 ± 0.23 |
| RMSD from the mean micelle initial structure (Å)† | 3.51 ± 0.26/3.21 ± 0.21 |
The flexible interface comprises segments Glu-1351–Gln-1356 and Gly-1364–Arg-1371 for EEA1 FYVE and Tyr-26, Val-27, Tyr-29, Arg-41–Glu-44, Glu-66–Tyr-79, Met-84, and Arg-88 for Vam7p PX.
Protein/micelle structures used as input for docking.
Spherical coordinates of the micelle center in the coordinate frame defined by the protein principal axes of inertia.
After superimposing the protein backbones.
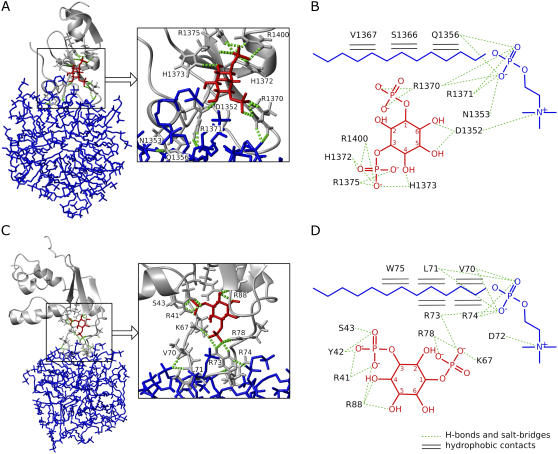
The protein-micelle interfaces of the best 20 models display a network of conserved hydrogen bonds to phosphate groups, salt bridges to choline amines, and hydrophobic contacts to the acyl chains of the DPC molecules (Fig. 4, A and B). Conserved intermolecular hydrogen bonds are found to involve Asn-1353, Gln-1356, Arg-1370, and Arg-1371, whereas the acidic residue Asp-1352 mediates salt bridges to the quaternary saturated amines of the choline groups. In addition to this electrostatic network, several hydrophobic contacts are found between the methyl and methylene groups of Gln-1356, Ser-1366, and Val-1367 and the acyl tails of DPC molecules (see Supplementary Table S1 for statistics). Together, this provides the first illustration, to our knowledge, of the ensemble of intermolecular interactions experienced by a FYVE domain upon membrane insertion.
FIGURE 4.
Intermolecular interfaces of the FYVE-DPC and PX-DPC complexes. The intermolecular hydrogen bonds present in the lowest energy structure of FYVE-DPC and PX-DPC complexes are shown in A and C, and the intermolecular interactions present in >30% of the best 20 structures are shown in B and D. The three-dimensional images depict proteins as gray ribbons, hydrogen-bonding residues as gray sticks, Ins(1,3)P2 in red, micelles in blue, and intermolecular hydrogen bonds as green dotted lines. The two-dimensional representations show DPC and Ins(1,3)P2 chemical structures together with the consensus intermolecular contacts to different protein residues (see also Supplementary Tables S1 and S2). Hydrogen bonds and salt bridges are drawn as green dotted lines, and the intermolecular hydrophobic contacts as black horizontal double lines.
The conserved intermolecular contacts represent a unified picture of the energetically favorable intermolecular interactions sampled by both phosphocholine and inositol binding determinants. It should be noted that the Ins(1,3)P2 binding pocket and its original hydrogen-bonding network were determined by distance restraints having the same weight as the protein-micelle interaction restraints. All these hydrogen bonds were found to have been preserved upon protein-micelle docking. An additional direct hydrogen bond involving the Arg-1400 side chain was discovered, suggesting more intimate lipid association than in the initial crystal structure (34). Two residues, Arg-1370 and Asp-1352, were predicted by the model to be dual determinants for both Ins(1,3)P2 binding and protein-micelle interaction, another new result which is consistent with the site-directed mutagenesis data. That is, the R1370A mutation causes a drastic decrease in the PtdIns(3)P binding affinity and disrupts the early endosome localization of the protein (46), whereas D1352A mutation results in a twofold decrease of Ins(1,3)P2 binding affinity (34), and the D1352V mutation causes partial cytoplasmic localization of the protein (47).
The micelle complex also explains further puzzles, such as why the V1369K mutation causes an enhancement of the endosomal targeting of the FYVE domain (48). The aliphatic side chain of Val-1369 is not involved in hydrophobic contacts with the hydrocarbon core but is rather located around the DPC phosphate groups; and therefore lysine substitution is likely to promote hydrogen bonds with the phospholipid headgroups. The residue that shows the largest number of hydrophobic contacts with the DPC acyl chain is Val-1367, in agreement with mutagenesis experiments in which the triple mutant S1366T-V1367F-V1369K showed a significant enhancement of the membrane affinity over the V1369K mutant, and the double mutant S1366T-V1369K had a similar membrane-targeting capability (48). In addition to the hydrophobic insertion of Val-1367, the side chain of the neighboring Thr-1368 hydrogen bonds to phosphate groups in 5 out of the 20 models, which explains why FYVE domains with double mutations V1367E-T1368E and V1367G-T1368G do not localize to early endosomes (47).
The FYVE domain's insertion angle defines its position relative to the micelle surface. This angle is defined by the principal inertia axis of the protein, which provides the minimal moment of inertia (the long inertia axis) and the vector connecting the protein with micelle centers. The insertion angle was determined from the DPC micelle complex to be 24.7° ± 11.4°. This angle is slightly smaller than the estimate of 48° ± 14° based on rigid body fitting of PRE data to a sphere (29) and compares well with the insertion angle of 26° ± 20° determined in the Orientation of Proteins in Membranes (OPM) database for optimal desolvation upon embedding the protein in a hydrophobic slab (49). Due to the different geometries of the membrane models, OPM insertion angles can only be directly compared to those from this docking procedure when the insertion angles are small and the distances between protein and micelle centers short. In addition, the protein-micelle geometries from the cross-validated runs compared well with each other and the fully restrained models, indicating that the angles were overdetermined (see Supplementary Table S3). A summary of the correspondence between the HADDOCK-derived protein-micelle structures and the NMR restraints, mutations that perturb the membrane interactions and the OPM-based protein-membrane models of EEA1 FYVE domain, is presented in Supplementary Table S4.
PX-DPC complex
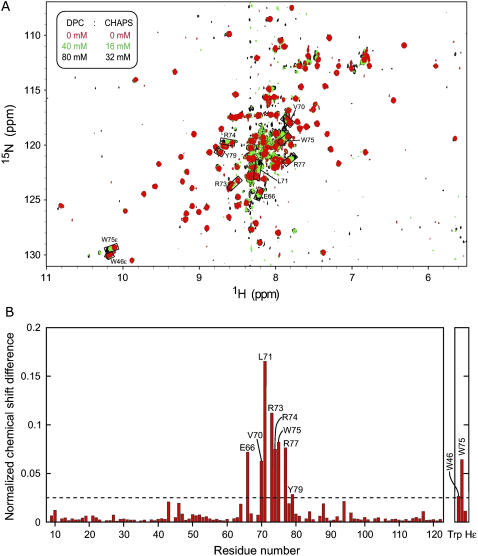
The micelle complex of the Vam7p PX domain, for which a solution structure of the free state is available (35), was modeled by applying the same strategy, albeit with fewer intermolecular restraints (Table 1). Three residues (Val-70, Leu-71, and Trp-75) showed PREs upon binding to spin-labeled diheptanoylphosphocoline (DHPC) micelles as well as large chemical shift perturbations, although there was no intermolecular NOE data available (28). The three hydrophobic residues form the tip of a solvent-accessible flexible loop (50), which is predicted to interact with the hydrocarbon region of a membrane by desolvation energy calculations in OPM. Therefore, they were restrained as deeply inserted residues (Fig. 1 B). Substantial chemical shift changes upon micelle addition (but not paramagnetic enhancements) were observed for the entire loop between Glu-66 and Tyr-79, suggesting conformational changes. Consequently, the whole loop was left flexible during the protein-micelle docking. In addition, residues found within 5 Å of the Ins(1,3)P2 ligand, including Tyr-26, Val-27, Tyr-29, Arg-41, Glu-44, Met-84, and Arg-88, were also left flexible to accommodate any structural changes induced in the binding pocket during protein-micelle complex formation. The ambiguous hydrogen-bond restraints used to drive the Ins(1,3)P2 docking (see the Methods section) were preserved during the protein-micelle calculations. Docking was performed on DPC micelles since they induce a similar pattern of chemical shift perturbations as DHPC (Fig. 5), indicating a similar mode of association with either micelle.
FIGURE 5.
Chemical shift changes of Vam7p PX amides upon DPC/CHAPS (3-[(3-cholamidopropyl)dimethylammonio]propane-1-sulfonic acid) titration. A shows three superimposed [1H,15N]HSQC spectra of Vam7p PX (65 μM) collected at 298 K on a Varian (Palo Alto, CA) INOVA 800 MHz spectrometer during titration with mixed DPC and CHAPS micelles (50 mM phosphate buffer, pH 7.0, 100 mM KCl, 1 mM Tris-(2-carboxyethyl) phosphine, and 1 mM NaN3). The histogram from B displays the normalized chemical shift changes (51) induced by the protein-micelle interactions. Residues with significant chemical shift changes are labeled.
The structural statistics of the energetically best 20 models of the 200 PX-micelle structures obtained after flexible docking and water refinement (Fig. 3 B) are presented in Table 2. The average RMSD of the protein interface is 1.34 ± 0.27 Å, the average deviation of the micelle centers from the mean is 6.5 ± 3.6 Å, and the angle between the protein long axis of inertia and the vector connecting the protein and micelle centers (insertion angle) is 38.7° ± 6.4°. The intermolecular nonbonded energies of the complex as well as the buried surface areas compare favorably to the FYVE domain values, in agreement with experimental results, which show similar micelle binding affinities for the two proteins (27,28).
The PX-micelle interfaces show an extensive network of hydrogen bonds to phosphate groups, salt bridges to choline amines, and hydrophobic contacts to the dodecyl chains that are supported by Vam7p mutagenesis experiments (Fig. 4, C and D, and Supplementary Table S2). Conserved intermolecular hydrogen bonds engage Val-70, Leu-71, Arg-73, and Arg-74 residues, whereas Asp-72 forms a salt bridge, and an array of hydrophobic contacts with the micelle core is evident for the Val-70, Leu-71, Arg-73, Arg-74, and Trp-75 residues. The Arg-73 residue is close to the tip of the membrane insertion loop and appears to be involved in both Ins(1,3)P2 and micelle binding, as suggested by the dual hydrogen-bonding network and hydrophobic contacts present in >30% of the 20 best structures. The critical role of Arg-73 is supported by the significantly reduced membrane binding and penetration capabilities of Vam7p PX upon its substitution with alanine in liposome binding and monolayer penetration experiments (28). The multitude of hydrophobic contacts observed in the complex implies their critical roles in membrane targeting, consistent with mutagenesis, liposome binding, and in vivo fluorescence microscopy experiments in which V70A–L71A and V70A-L71A-W75A mutants failed to localize to liposomes and yeast vacuole membranes (28). Additionally, monolayer surface tension experiments demonstrated that V70A, L71A, and W75A point mutations reduce the membrane-penetrating ability of the Vam7p PX domain. The strongest reduction was observed for Leu-71, which is the residue which shows the largest number of hydrophobic contacts in the micelle complexes.
Novel interactions can be seen in the Vam7p PX-micelle complex. Docking models predict for the first time, to our knowledge, that residues Asp-72 and Arg-74 are determinants for DPC micelle binding and reveal novel Ins(1,3)P2 contacts for Tyr-42, Ser-43, and Lys-67 as well as the bivalent role of Arg-73 for inositol and phosphocholine interactions. The interaction between the 1-phosphate group of the inositol ligand and the side chain of Lys-67 is highly conserved among PX domains and has been shown to be essential for PtdIns(3)P recognition in p40phox and Grd19p PX. The alanine mutation of the corresponding residue of p40phox, Lys-92, resulted in a drastic reduction of the protein affinity for PtdIns(3)P, whereas the mutation of Arg-60, another 1-phosphate hydrogen-bonding residue, caused only a moderate decrease in affinity (42). In Grd19p PX, the residue corresponding to Vam7p Lys-67 (Lys-112) is the key factor in the conformational change of the adjacent residues upon PtdIns(3)P binding (43). Therefore, the docking result not only provides the structural basis of intermolecular interactions responsible for Vam7p association with membrane-mimicking micelles but also reveals the conserved determinants of PX:PtdIns(3)P headgroup recognition.
The protein-micelle docking methodology presented here enables the structural characterization of proteins complexed with membrane models based on experimental evidence for membrane interactions. Two peripheral membrane proteins which are paradigms for phosphoinositide recognition and endocytic membrane targeting, the EEA1 FYVE and Vam7p PX domains, were characterized to show how protein recruitment to membranes involves a network of specific and nonspecific interactions to an array of lipid groups within the bilayer matrix. The synergy of these multiple interactions contributes to the overall affinity and specificity required for efficient membrane docking. The structural results provide a mechanistic explanation for diverse experimental data regarding the effects of mutations and posttranslational modifications on phospholipid recognition and membrane targeting of peripheral membrane proteins.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank S. Rajesh for help with sample preparations, C. Ludwig for discussions and help with relaxation measurements, and the Henry Wellcome Building for Biomolecular NMR Spectroscopy for access to NMR and computational facilities.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council, European Union, and Wellcome Trust.
Editor: Arthur G. Palmer III.
References
- 1.Cho, W., and R. V. Stahelin. 2005. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 34:119–151. [DOI] [PubMed] [Google Scholar]
- 2.Lin, Y., R. Nielsen, D. Murray, W. L. Hubbell, C. Mailer, B. H. Robinson, and M. H. Gelb. 1998. Docking phospholipase A2 on membranes using electrostatic potential-modulated spin relaxation magnetic resonance. Science. 279:1925–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frazier, A. A., M. A. Wisner, N. J. Malmberg, K. G. Victor, G. E. Fanucci, E. A. Nalefski, J. J. Falke, and D. S. Cafiso. 2002. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 41:6282–6292. [DOI] [PubMed] [Google Scholar]
- 4.Kohout, S. C., S. Corbalan-Garcia, J. C. Gomez-Fernandez, and J. J. Falke. 2003. C2 domain of protein kinase C alpha: elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry. 42:1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malmberg, N. J., and J. J. Falke. 2005. Use of EPR power saturation to analyze the membrane-docking geometries of peripheral proteins: applications to C2 domains. Annu. Rev. Biophys. Biomol. Struct. 34:71–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, E. R., and A. F. Davis. 1998. Direct interaction of a Ca2+-binding loop of synaptotagmin with lipid bilayers. J. Biol. Chem. 273:13995–14001. [DOI] [PubMed] [Google Scholar]
- 7.Perisic, O., H. F. Paterson, G. Mosedale, S. Lara-Gonzalez, and R. L. Williams. 1999. Mapping the phospholipid-binding surface and translocation determinants of the C2 domain from cytosolic phospholipase A2. J. Biol. Chem. 274:14979–14987. [DOI] [PubMed] [Google Scholar]
- 8.Medkova, M., and W. Cho. 1999. Interplay of C1 and C2 domains of protein kinase C-alpha in its membrane binding and activation. J. Biol. Chem. 274:19852–19861. [DOI] [PubMed] [Google Scholar]
- 9.Stahelin, R. V., F. Long, K. Diraviyam, K. S. Bruzik, D. Murray, and W. Cho. 2002. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J. Biol. Chem. 277:26379–26388. [DOI] [PubMed] [Google Scholar]
- 10.Stahelin, R. V., A. Burian, K. S. Bruzik, D. Murray, and W. Cho. 2003. Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J. Biol. Chem. 278:14469–14479. [DOI] [PubMed] [Google Scholar]
- 11.Malkova, S., F. Long, R. V. Stahelin, S. V. Pingali, D. Murray, W. Cho, and M. L. Schlossman. 2005. X-ray reflectivity studies of cPLA2(alpha)-C2 domains adsorbed onto Langmuir monolayers of SOPC. Biophys. J. 89:1861–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malkova, S., R. V. Stahelin, S. V. Pingali, W. Cho, and M. L. Schlossman. 2006. Orientation and penetration depth of monolayer-bound p40phox-PX. Biochemistry. 45:13566–13575. [DOI] [PubMed] [Google Scholar]
- 13.Allegrini, P. R., G. J. van Scharrenburg, A. J. Slotboom, G. H. de Haas, and J. Seelig. 1985. Side-chain dynamics of two aromatic amino acids in pancreatic phospholipase A2 as studied by deuterium nuclear magnetic resonance. Biochemistry. 24:3268–3273. [DOI] [PubMed] [Google Scholar]
- 14.Tuzi, S., N. Uekama, M. Okada, S. Yamaguchi, H. Saito, and H. Yagisawa. 2003. Structure and dynamics of the phospholipase C-delta1 pleckstrin homology domain located at the lipid bilayer surface. J. Biol. Chem. 278:28019–28025. [DOI] [PubMed] [Google Scholar]
- 15.Roberts, M. F., and A. G. Redfield. 2004. High-resolution 31p field cycling NMR as a probe of phospholipid dynamics. J. Am. Chem. Soc. 126:13765–13777. [DOI] [PubMed] [Google Scholar]
- 16.Vogel, A., K. T. Tan, H. Waldmann, S. E. Feller, M. F. Brown, and D. Huster. 2007. Flexibility of Ras lipid modifications studied by 2H solid-state NMR and molecular dynamics simulations. Biophys. J. 93:2697–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohda, D., and F. Inagaki. 1992. Structure of epidermal growth factor bound to perdeuterated dodecylphosphocholine micelles determined by two-dimensional NMR and simulated annealing calculations. Biochemistry. 31:677–685. [DOI] [PubMed] [Google Scholar]
- 18.Kutateladze, T., and M. Overduin. 2001. Structural mechanism of endosome docking by the FYVE domain. Science. 291:1793–1796. [DOI] [PubMed] [Google Scholar]
- 19.Zamoon, J., A. Mascioni, D. D. Thomas, and G. Veglia. 2003. NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys. J. 85:2589–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Rio, A., K. Dutta, J. Chavez, I. Ubarretxena-Belandia, and R. Ghose. 2007. Solution structure and dynamics of the N-terminal cytosolic domain of rhomboid intramembrane protease from Pseudomonas aeruginosa: insights into a functional role in intramembrane proteolysis. J. Mol. Biol. 365:109–122. [DOI] [PubMed] [Google Scholar]
- 21.Jaud, S., D. J. Tobias, J. J. Falke, and S. H. White. 2007. Self-induced docking site of a deeply embedded peripheral membrane protein. Biophys. J. 92:517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hritz, J., J. Ulicny, A. Laaksonen, D. Jancura, and P. Miskovsky. 2004. Molecular interaction model for the C1B domain of protein kinase C-gamma in the complex with its activator phorbol-12-myristate-13-acetate in water solution and lipid bilayer. J. Med. Chem. 47:6547–6555. [DOI] [PubMed] [Google Scholar]
- 23.Xu, G. Y., T. McDonagh, H. A. Yu, E. A. Nalefski, J. D. Clark, and D. A. Cumming. 1998. Solution structure and membrane interactions of the C2 domain of cytosolic phospholipase A2. J. Mol. Biol. 280:485–500. [DOI] [PubMed] [Google Scholar]
- 24.Wang, G., J. T. Sparrow, and R. J. Cushley. 1997. The helix-hinge-helix structural motif in human apolipoprotein A-I determined by NMR spectroscopy. Biochemistry. 36:13657–13666. [DOI] [PubMed] [Google Scholar]
- 25.Papavoine, C. H., R. N. Konings, C. W. Hilbers, and F. J. van de Ven. 1994. Location of M13 coat protein in sodium dodecyl sulfate micelles as determined by NMR. Biochemistry. 33:12990–12997. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura, T., H. Takahashi, K. Takeuchi, T. Kohno, K. Wakamatsu, and I. Shimada. 2005. Direct determination of a membrane-peptide interface using the nuclear magnetic resonance cross-saturation method. Biophys. J. 89:4051–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutateladze, T. G., D. G. Capelluto, C. G. Ferguson, M. L. Cheever, A. G. Kutateladze, G. D. Prestwich, and M. Overduin. 2004. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 279:3050–3057. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. A., J. Kovacs, R. V. Stahelin, M. L. Cheever, M. Overduin, T. G. Setty, C. G. Burd, W. Cho, and T. G. Kutateladze. 2006. Molecular mechanism of membrane docking by the Vam7p PX domain. J. Biol. Chem. 281:37091–37101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunecky, R., S. Lee, P. W. Rzepecki, M. Overduin, G. D. Prestwich, A. G. Kutateladze, and T. G. Kutateladze. 2005. Investigation of the binding geometry of a peripheral membrane protein. Biochemistry. 44:16064–16071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solomon, I. 1955. Relaxation processes in a system of two spins. Phys. Rev. 99:559–565. [Google Scholar]
- 31.Iwahara, J., C. D. Schwieters, and G. M. Clore. 2004. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 126:5879–5896. [DOI] [PubMed] [Google Scholar]
- 32.Dominguez, C., R. Boelens, and A. M. Bonvin. 2003. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125:1731–1737. [DOI] [PubMed] [Google Scholar]
- 33.Rice, L. M., and A. T. Brunger. 1994. Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins. 19:277–290. [DOI] [PubMed] [Google Scholar]
- 34.Dumas, J. J., E. Merithew, E. Sudharshan, D. Rajamani, S. Hayes, D. Lawe, S. Corvera, and D. G. Lambright. 2001. Multivalent endosome targeting by homodimeric EEA1. Mol. Cell. 8:947–958. [DOI] [PubMed] [Google Scholar]
- 35.Lu, J., J. Garcia, I. Dulubova, T. C. Sudhof, and J. Rizo. 2002. Solution structure of the Vam7p PX domain. Biochemistry. 41:5956–5962. [DOI] [PubMed] [Google Scholar]
- 36.Tieleman, D. P., D. van der Spoel, and H. J. C. Berendsen. 2000. Molecular dynamics simulations of dodecylphosphocholine micelles at three different aggregate sizes: micellar structure and lipid chain relaxation. J. Phys. Chem. B. 104:6380–6388. [Google Scholar]
- 37.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921. [DOI] [PubMed] [Google Scholar]
- 38.Schuttelkopf, A. W., and D. M. van Aalten. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60:1355–1363. [DOI] [PubMed] [Google Scholar]
- 39.Linge, J. P., M. A. Williams, C. A. Spronk, A. M. Bonvin, and M. Nilges. 2003. Refinement of protein structures in explicit solvent. Proteins. 50:496–506. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen, W. L., and J. Tirado-Rives. 1988. The OPLS potential function for proteins. Energy minimization for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 110:1657–1666. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 42.Bravo, J., D. Karathanassis, C. M. Pacold, M. E. Pacold, C. D. Ellson, K. E. Anderson, P. J. Butler, I. Lavenir, O. Perisic, P. T. Hawkins, L. Stephens, and R. L. Williams. 2001. The crystal structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate. Mol. Cell. 8:829–839. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, C. Z., I. L. de La Sierra-Gallay, S. Quevillon-Cheruel, B. Collinet, P. Minard, K. Blondeau, G. Henckes, R. Aufrere, N. Leulliot, M. Graille, I. Sorel, P. Savarin, F. de la Torre, A. Poupon, J. Janin, and H. van Tilbeurgh. 2003. Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J. Biol. Chem. 278:50371–50376. [DOI] [PubMed] [Google Scholar]
- 44.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- 45.Hubbard, S. J., and J. M. Thornton. 1993. ‘NACCESS’, Computer Program. Department of Biochemistry and Molecular Biology, University College London, UK.
- 46.Gaullier, J. M., E. Ronning, D. J. Gillooly, and H. Stenmark. 2000. Interaction of the EEA1 FYVE finger with phosphatidylinositol 3-phosphate and early endosomes. Role of conserved residues. J. Biol. Chem. 275:24595–24600. [DOI] [PubMed] [Google Scholar]
- 47.Kutateladze, T. G., K. D. Ogburn, W. T. Watson, T. de Beer, S. D. Emr, C. G. Burd, and M. Overduin. 1999. Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol. Cell. 3:805–811. [DOI] [PubMed] [Google Scholar]
- 48.Hayakawa, A., S. J. Hayes, D. C. Lawe, E. Sudharshan, R. Tuft, K. Fogarty, D. Lambright, and S. Corvera. 2004. Structural basis for endosomal targeting by FYVE domains. J. Biol. Chem. 279:5958–5966. [DOI] [PubMed] [Google Scholar]
- 49.Lomize, M. A., A. L. Lomize, I. D. Pogozheva, and H. I. Mosberg. 2006. OPM: orientations of proteins in membranes database. Bioinformatics. 22:623–625. [DOI] [PubMed] [Google Scholar]
- 50.Cheever, M. L., T. G. Kutateladze, and M. Overduin. 2006. Increased mobility in the membrane targeting PX domain induced by phosphatidylinositol 3-phosphate. Protein Sci. 15:1873–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grzesiek, S., S. J. Stahl, P. T. Wingfield, and A. Bax. 1996. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 35:10256–10261. [DOI] [PubMed] [Google Scholar]