Abstract
Aquaporin-1 (AQP1) is an integral membrane protein that facilitates osmotic water transport across cell plasma membranes in epithelia and endothelia. AQP1 has no known specific interactions with cytoplasmic or membrane proteins, but its recovery in a detergent-insoluble membrane fraction has suggested possible raft association. We tracked the membrane diffusion of AQP1 molecules labeled with quantum dots at an engineered external epitope at frame rates up to 91 Hz and over times up to 6 min. In transfected COS-7 cells, >75% of AQP1 molecules diffused freely over ∼7 μm in 5 min, with diffusion coefficient, D1-3 ∼ 9 × 10−10 cm2/s. In MDCK cells, ∼60% of AQP1 diffused freely, with D1-3 ∼ 3 × 10−10 cm2/s. The determinants of AQP1 diffusion were investigated by measurements of AQP1 diffusion following skeletal disruption (latrunculin B), lipid/raft perturbations (cyclodextrin and sphingomyelinase), and bleb formation. We found that cytoskeletal disruption had no effect on AQP1 diffusion in the plasma membrane, but that diffusion was increased greater than fourfold in protein de-enriched blebs. Cholesterol depletion in MDCK cells greatly restricted AQP1 diffusion, consistent with the formation of a network of solid-like barriers in the membrane. These results establish the nature and determinants of AQP1 diffusion in cell plasma membranes and demonstrate long-range nonanomalous diffusion of AQP1, challenging the prevailing view of universally anomalous diffusion of integral membrane proteins, and providing evidence against the accumulation of AQP1 in lipid rafts.
INTRODUCTION
The aquaporins (AQPs) are a family of small, integral membrane proteins whose primary function is to facilitate osmotically driven water transport across cell plasma membranes. There are at least 10 AQP isoforms in mammals and many more in lower organisms and plants. One isoform, AQP1, is expressed widely in epithelial and endothelial cells, where it plays an important role in the urinary concentrating system, fluid secretion in the eye and brain, and angiogenesis (reviewed in Verkman (1)). AQP1 monomers (molecular weight ∼30,000), which consist of six membrane-spanning α-helical segments forming a twisted barrel, are assembled in membranes as stable tetramers with individual AQP1 monomers containing aqueous pores (2–4).
AQP1 is constitutively active and targeted to the cell plasma membrane. In kidney epithelium AQP1 is present in both the apical and basolateral membranes (5–7), whereas it is found only in the apical membrane in epithelial cells of choroid plexus in the central nervous system (8). The N-glycosylation site on AQP1 may facilitate its targeting to the apical plasma membrane, a process that has been linked to lipid rafts (9). Following cold Triton X-100 extraction of brain and lung cells, AQP1 has been found in fractions containing putative raft-associated molecules such as the ganglioside GM1, sphingomyelin, cholesterol, flottilin, and caveolin; however, immunoblotting of detergent-insoluble AQP1 showed both glycosylated and nonglycosylated forms (10,11).
To avoid the caveats of biochemical assays based on detergent solubility as a measure of lipid-raft association, we measured the diffusion of AQP1 in plasma membranes of living cells to investigate its possible confinement within microdomains. The question of raft association of lipids and proteins has been addressed by diffusion measurements using fluorescence recovery after photobleaching and single particle tracking (SPT) (12–17). Two photobleaching studies have previously investigated the diffusion of AQP1 in cell membranes. Using an FITC-labeled antibody against a glycocalyx epitope on erythrocyte AQP1, Cho et al. (18) reported moderately slowed AQP1 diffusion with a significant immobile fraction (30–40%). Notwithstanding caveats about possible antibody-induced protein cross-linking and the limited spatial resolution afforded by the small erythrocyte size, it was concluded from mechanical deformation studies that AQP1 mobility was restricted by the spectrin membrane skeleton by direct interactions and/or spectrin-dependent alterations in membrane structure. Our lab used functional GFP-AQP chimeras expressed in mammalian kidney cells to study AQP1 diffusion by spot and image photobleaching (19). In contrast to the data of Cho et al. (18), AQP1 diffusion was substantially faster and AQP1 appeared to be fully mobile. Photobleaching measurements on a GFP-AQP2 chimera showed slower and cAMP-regulated mobility that involved AQP2 interactions with the actin skeleton (19). GFP-AQP2 diffusion was substantially faster at the endoplasmic reticulum (20). However, because of the intrinsic limitation in photobleaching of ensemble averaging, it was not possible to resolve the nature and determinants of the mobility of individual AQP tetramers.
The purpose of this study was to characterize at high resolution the motion of individual AQP1 tetramers in mammalian cell plasma membranes. For these measurements we tracked the movement of AQP1 after labeling with quantum dots (Qdots) at an engineered external c-myc epitope. The c-myc tag in the AQP1 construct was shown previously not to interfere with AQP1 function or cellular processing (21). SPT measurements were done at frame rates up to 91 Hz and acquisitions times up to 6 min to address the biophysical nature and determinants of AQP1 diffusion. We used COS-7 cells, a nonpolarized fibroblast, and Madine-Darby canine kidney (MDCK) cells, an epithelial cell line that has been studied extensively in the fields of lipid rafts and aquaporins (22,23). Under control conditions, we found little evidence of long-term confinement of AQP1 within membrane microdomains caused by lipid or cytoskeletal barriers. Instead, we found largely simple (nonanomalous) Brownian diffusion over distances up to 18 μm in 5 min. Several other interesting observations were made, including remarkable variation in AQP1 diffusion coefficients in different cell types, and exceptionally rapid AQP1 diffusion in relatively protein-free membrane blebs.
METHODS
Cell culture and transfections
Plasmid pSP64.AQP1.T120.myc, which encodes human AQP1 containing a c-myc sequence inserted between residues T120 and G121 (21), was ligated into mammalian expression vector pcDNA3.1 (Invitrogen, Carlsbad, CA) at EcoRI and XbaI restriction sites, and the sequence confirmed. Cell lines included COS-7 cells (ATCC No. CRL-1651), MDCK cells (ATCC No. CCL-34), and MDCK type II cells (MDCK II, ECACC No. 00062107). All cell cultures were maintained at 37°C in 5% CO2 / 95% air in DMEM-H21 medium containing 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin. Six hours before transfection, cells were plated in 12-well plates containing 18-mm diameter glass coverslips using antibiotic-free medium. Cells were transfected 24–48 h before experiments with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Labeling and cell treatments
Labeling was done on confluent cell monolayers. Before labeling of AQP1 with Qdots, cells were washed with 3 mL of phosphate-buffered saline containing 6 mM glucose and 1 mM pyruvate (GP buffer) and incubated for 5 min in blocking buffer (GP buffer containing 1% bovine serum albumin), followed by 10 min with 0.1 μg/mL mouse anti-c-myc antibody (Roche Applied Science, Indianapolis, IN) in blocking buffer. Cells were then rinsed 5 times with GP buffer and incubated for 5 min with 0.1 nM goat F(ab′)2 anti-mouse IgG-conjugated Qdots (Invitrogen) in blocking buffer. For labeling of plasma membrane lipids with Qdots, 1 μL of a 0.1 mg/mL ethanol solution of N-biotinyl dipalmitoyl phosphatidylethanolamine (bi-DPPE; Avanti Polar Lipids, Alabaster, AL) was injected into 1 mL of GP buffer over cells and incubated for 20 min at 37°C. Excess bi-DPPE was washed off and cells were incubated with blocking buffer for 5 min, followed by 5 min with 0.1 nM streptavidin-conjugated Qdots in blocking buffer. For plasma membrane labeling with fluorescent lipids, 10 μL of a 1 mg/mL ethanol solution of N-(lissamine rhodamine B sulphonyl) dipalmitoyl phosphatidylethanolamine ((Rh-DPPE); Avanti) was injected into 1 mL of GP buffer at room temperature. Cells were incubated with the lipid suspension for 10 min at 37°C. Immediately following the above labeling procedures, cells were rinsed 5 times with GP buffer, coverslips were transferred to a custom-built perfusion chamber, and maintained in GP buffer throughout the experiment.
For disruption of the cytoskeleton, cells were incubated with 0.5 μM latrunculin B (Invitrogen) in GP buffer at 37°C for 30 min before labeling. Latrunculin was also included in the labeling and experimental bathing solutions. In some experiments plasma membrane lipid composition was modified by 30 min incubation in GP buffer at 37°C with 10 mM methyl-β-cyclodextrin ((CD); Sigma, St. Louis, MO) or 20 min incubation with 0.05 units/mL sphingomyelinase ((SMase); Sigma). These treatments have been shown to reduce cellular cholesterol or sphingomyelin by ∼50%, respectively (24,25). In some studies, cells were fixed just after labeling by 20 min incubation with PBS containing 4% paraformaldehyde.
SPT instrumentation and measurements
SPT and imaging of GFP- and Qdot-labeled cells was performed using a Nikon Eclipse TE2000U inverted epifluorescence microscope (Nikon, Melville, NY) equipped with an Exfo X-Cite light source (Exfo, Mississauga, Ontario, Canada) and computer-controlled shutter (Uniblitz; Vincent Associates, Rochester, NY). Cells labeled with Qdots were imaged through a Nikon 100× TIRF oil immersion objective (numerical aperture 1.45). Qdot fluorescence was excited using a 420/40× excitation filter and 470DCXR dichroic mirror, and detected through a 655/40m emission filter (Chroma, Rockingham, VT). GFP fluorescence was excited and detected through the same 100× objective and Chroma filter set No. 31001. Images were acquired by a Hamamatsu EM-CCD deep-cooled camera (Hamamatsu, Bridgewater, NJ). Data were obtained at 37°C within 30 min of the final wash step after cell labeling. Long-range SPT was done using time-lapse acquisition in which 100-ms exposures were acquired at a rate of 1 Hz for 6 min, with the illumination light shuttered between exposures. Short-range SPT was done using continuous 11-ms acquisitions for 6 s (91 Hz), or 31-ms acquisitions for 20 s (32 Hz). The spatial resolution of the system, determined from the standard deviation of x,y-coordinates of immobilized Qdots (26), was 18 nm at our fastest frame rate. Image sequences were analyzed and trajectories constructed using IDL software (Research Systems, Boulder, CO) with algorithms available as shareware at http://www.physics.emory.edu/faculty/weeks/. For long-range time-lapse SPT, only trajectories longer than 60 steps were analyzed, whereas only trajectories longer than 100 steps were analyzed from continuous short-range SPT. Blinking of individual Qdots was accounted for during trajectory constructions, and included in subsequent calculations. Trajectories were analyzed as described below using custom programs written in LabVIEW (National Instruments, Austin, TX).
SPT analysis
For each trajectory, the mean-squared displacement (MSD), 〈r2(t)〉 was calculated as:
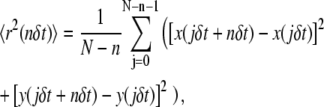 |
(1) |
where δt is the temporal resolution of the acquisition, x(jδt), y(jδt) is the particle coordinate at time t = jδt, N is the total number of frames recorded for an individual particle, and n = 0, 1, 2,…, N − 1. A linear least-squares fit to the first three points of the MSD versus t-curve, 〈r2(t)〉1-3, was done to compute the microscopic diffusion coefficient, D1-3, and the “offset” resulting from uncertainty in the position of the centroid of the particle:
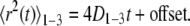 |
(2) |
The offset was subtracted from each point on the MSD versus t-curve, and the first 25% of the curve (27) was fitted using a weighted Levenberg-Marquardt nonlinear least-squares fitting algorithm to a combined quadratic, polynomial, and exponential function with fitting parameters a1, a2, a3 ≥ 0 (28):
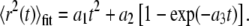 |
(3) |
The fit was weighted by the variance in the MSD at each time step. The range of an individual particle at specific time t was then computed as:
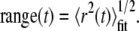 |
(4) |
The relative deviation (RD) (29) of the MSD from linearity, measured at the nth step of a trajectory with total length N, was defined as:
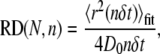 |
(5) |
where 4D0 is the initial slope of 〈r2(t)〉fit. Immobile particles were defined as those with a range <18 nm (limit of positional accuracy) after 100 steps. Mean values of D1-3 and range reported in the text and Table 1 were derived from mobile particles only. Statistical significance of differences in mean values was determined using the Student's t-test. Pooled MSD data shown in Figs. 1 D and 2 B were averaged from mobile particles only, whereas cumulative probability plots (Fig. 2, C and D) were compiled for all particles, mobile and immobile.
TABLE 1.
Diffusion of free and restricted fractions of AQP1 following various treatments
| Free
|
Restricted
|
Free + Restricted
|
|||||
|---|---|---|---|---|---|---|---|
| D1-3 (μm2/s)* | Range† (μm)* | D1-3 (μm2/s)* | Range† (μm)* | D1-3 (μm2/s)* | Range† (μm)* | ||
| COS-7 | |||||||
| Control‡ | 751 (33) | 0.089 ± 0.006 | 0.53 ± 0.02 | 0.072 ± 0.012 | 0.24 ± 0.02 | 0.084 ± 0.006 | 0.47 ± 0.02 |
| + latrunculin | 604 (22) | 0.083 ± 0.006 | 0.50 ± 0.02 | 0.061 ± 0.008 | 0.23 ± 0.02 | 0.077 ± 0.006 | 0.44 ± 0.02 |
| + CD | 559 (68) | 0.054 ± 0.003§ | 0.38 ± 0.01§ | 0.051 ± 0.005 | 0.19 ± 0.01 | 0.055 ± 0.003§ | 0.32 ± 0.01§ |
| + SMase | 267 (42) | 0.093 ± 0.008 | 0.54 ± 0.03 | 0.067 ± 0.011 | 0.20 ± 0.02 | 0.089 ± 0.008 | 0.45 ± 0.03 |
| MDCK | |||||||
| Control‡ | 763 (47) | 0.026 ± 0.003 | 0.25 ± 0.02 | 0.020 ± 0.003 | 0.113 ± 0.007 | 0.025 ± 0.003 | 0.21 ± 0.02 |
| + latrunculin | 703 (50) | 0.037 ± 0.005 | 0.29 ± 0.02 | 0.030 ± 0.005 | 0.14 ± 0.01 | 0.036 ± 0.005 | 0.24 ± 0.02 |
| + CD | 253 (18) | 0.006 ± 0.001§ | 0.10 ± 0.01§ | 0.011 ± 0.002 | 0.054 ± 0.005§ | 0.011 ± 0.001§ | 0.067 ± 0.008§ |
| + SMase | 287 (24) | 0.06 ± 0.02 | 0.25 ± 0.05 | 0.040 ± 0.009 | 0.12 ± 0.01 | 0.044 ± 0.010 | 0.16 ± 0.02 |
| MDCK II | |||||||
| Control‡ | 702 (24) | 0.037 ± 0.003 | 0.32 ± 0.02 | 0.027 ± 0.002 | 0.143 ± 0.009 | 0.034 ± 0.003 | 0.26 ± 0.01 |
| + latrunculin | 408 (24) | 0.051 ± 0.004§ | 0.40 ± 0.02 | 0.036 ± 0.004 | 0.17 ± 0.01 | 0.046 ± 0.004 | 0.33 ± 0.02 |
| + CD | 329 (15) | 0.011 ± 0.004§ | 0.14 ± 0.03§ | 0.016 ± 0.001§ | 0.064 ± 0.005§ | 0.015 ± 0.002§ | 0.08 ± 0.01§ |
| + SMase | 187 (15) | 0.15 ± 0.03§ | 0.53 ± 0.07§ | 0.10 ± 0.03§ | 0.20 ± 0.03 | 0.12 ± 0.02§ | 0.37 ± 0.05 |
Mean ± SE.
Range at 1 s.
Total number of trajectories, with number of cells in parentheses.
P < 0.01 when compared to untreated cells.
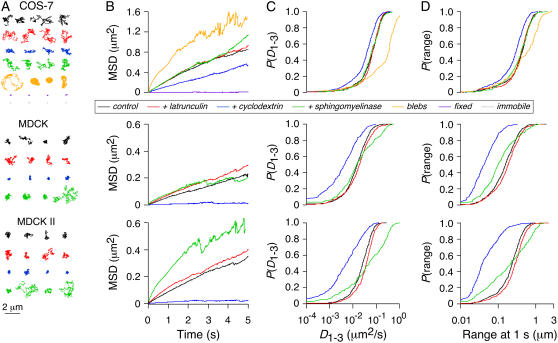
FIGURE 1.
Labeling and time-lapse SPT of AQP1 in the plasma membrane. (A) Schematic of Qdot-labeled AQP1 monomer. Human c-myc epitope was inserted between residues T120 and G121 (red), which reside in the second extracellular loop (yellow), between transmembrane helices M4 and M5 (green). This site is well removed from the two NPA motifs (blue) that serve as the selectivity filter of the pore. (B) Labeling of AQP1.myc with high density of Qdots on the surface of live COS-7 (left) and MDCK (right) cells. (C) MDCK cells cotransfected with cytoplasmic GFP (green) and AQP1.myc, labeled at low density (as used for SPT measurements) with Qdots (red). Arrow points to cell expressing GFP without AQP1.myc. (D) Examples of trajectories from time-lapse SPT acquired at 1 Hz (total time, 6 min) overlaid onto fluorescence images of GFP in the cytoplasm of a COS-7 cell (left) and a MDCK cell (right). (E) Mean-squared displacement (MSD) versus time curves from all time-lapse SPT (∼300 individual trajectories averaged for each cell type).
FIGURE 2.
SPT analysis of AQP1 at high time resolution (91 Hz). (A) Representative trajectories for AQP1 diffusing in the plasma membrane of COS-7 (top), MDCK (middle), and MDCK II (bottom) cells shown under control conditions (black), and after treatment with latrunculin (red), cyclodextrin (blue), or sphingomyelinase (green). For COS-7 cells, trajectories are also shown for paraformaldehyde-fixed cells (purple) and on the surface of membrane blebs (orange). Trajectories for Qdots immobilized on glass are shown for comparison (gray). (B) Corresponding combined MSD versus time curves for each cell type/maneuver shown. Cumulative probability distributions shown for AQP1 diffusion coefficient, D1-3 (C), and range at 1 s (D).
Photobleaching measurements of a fluorescent lipid
Cells were mounted on a Nikon inverted epifluorescence microscope in a custom-built chamber maintained at 37°C. An argon-ion laser beam (Innova I-308C; Coherent, Palo Alto, CA) was modulated by an acoustooptic modulator (Brimrose, Baltimore, MD) and directed onto the sample through a 60× oil immersion objective (Nikon, numerical aperture 1.40). A focused beam width of 1.4 μm at half-maximum was determined by measuring recoveries of control solutions with known diffusion coefficient. Laser excitation at 514 nm was reflected onto the sample by a 565-nm dichroic, with sample fluorescence observed through a 605/50 emission filter (Chroma). For cell visualization and selection of spot position before acquisition, full-field epifluorescence was accomplished by defocusing the undiffracted beam and redirecting it onto the sample through a fiber optic. Fluorescence was detected by a photomultiplier (Thorn-EMI, London, UK), digitized at 14-bit resolution, and collected using a custom program written in LabVIEW. A computer-controlled shutter (Uniblitz) illuminated the sample at a rate of 2 Hz. Prebleach and postbleach intensities were acquired as an average of 10,000 samples collected during the shutter open time of 20 ms. For each fluorescence recovery experiment, the apparent diffusion coefficient and mobile fraction was determined from pre- and postbleach intensities, and the half-time of recovery, as described previously (30,31).
RESULTS
Plasma membrane diffusion of Qdot-labeled AQP1
Following transfection, cells expressing AQP1.myc were labeled with Qdots as described under Methods. The c-myc epitope was inserted in an extracellular loop of AQP1 at a location that does not interfere with its function or processing (Fig. 1 A). Labeling was done on confluent cell monolayers, which limited labeling to the apical plasma membrane. Heavily labeled cells showed fairly uniform fluorescence over the entire apical membrane (Fig. 1 B). Qdot labeling was highly specific. Cells that did not express AQP1.myc generally showed no Qdots under labeling conditions used for SPT measurements in which AQP1.myc expressing cells showed multiple distinguishable Qdots (Fig. 1 C).
We initially characterized long-range motions of AQP1 in cell membranes using time-lapse image acquisition at 1 Hz over 6 min. Cells were cotransfected with GFP to readily identify cell edges to ensure that the observed diffusion was indeed on the cell membrane. Examples of Qdot trajectories are shown in Fig. 1 D (see also Supplementary Material, Movie 1). AQP1 diffused more rapidly and covered a much larger area over the 6-min acquisition in COS-7 than in MDCK cells. The range (Eq. 4) of AQP1 at 1 min was 2.0 ± 0.1 μm (mean ± SE, 325 trajectories, eight cells) in COS-7 cells, compared to 0.87 ± 0.07 μm (302 trajectories, 17 cells) in MDCK cells. The MSD versus time plots in both cell types were approximately linear over 5 min (Fig. 1 E). Although time-lapse acquisition at 1 Hz provided a good overview of the long-range motions of AQP1, a much faster rate of acquisition was necessary to characterize the modes of diffusion of individual molecules and to accurately determine diffusion coefficients.
A large series of SPT measurements was done with continuous image acquisition at 91 Hz over 6 s (546 frames), characterizing short-range diffusion in COS-7, MDCK, and MDCK II cells. In all three types of cells, <1% of observed Qdots were immobile. Fig. 2 shows representative trajectories (A), averaged MSD plots (B), and cumulative probability plots for diffusion coefficient D1-3 ((C); Eq. 2) and range at 1 s ((D); Eq. 4). AQP1 in COS-7 cells had an average D1-3 of 0.084 ± 0.006 μm2/s (751 trajectories, 33 cells), compared to a D1-3 of 0.025 ± 0.003 μm2/s (763 trajectories, 47 cells) in MDCK cells (Table 1). For comparison, we also measured the diffusion of a Qdot-linked lipid bi-DPPE in COS-7 cells (not shown). As expected, lipid diffusion was faster than that of AQP1, with an average D1-3 of 0.21 ± 0.01 μm2/s and range of 0.77 ± 0.03 μm at 1 s (421 trajectories, 30 cells).
Several types of measurements were done to explore diffusive mechanisms in COS-7 and MDCK cells, including analysis of effects of cytoskeletal and lipid/raft perturbation, diffusion measurements of AQP1 in relatively protein-depleted membrane blebs, and analysis of diffusive modes of individual trajectories.
For several membrane proteins, cytoskeletal interactions have been shown to be a determinant of their diffusion and confinement (32–34). A recent example characterized by our lab is the cystic fibrosis transmembrane conductance regulator protein, CFTR, in which F-actin disruption by latrunculin B greatly increased diffusion (28). However, unlike CFTR, AQP1 diffusion was not affected by latrunculin B in any of the cell types studied (Fig. 2; Table 1), indicating that AQP1 is not restricted by actin skeletal interactions.
AQP1 diffusion was also observed in membrane blebs, which are cytoskeleton-free, relatively protein de-enriched fragments of the cell plasma membrane that can appear spontaneously or after DMSO or other treatments (35,36). Blebs occasionally appeared on the surface of some COS-7 cells at 48–72 h after transfection, 6–12 h after they reached confluence. Trajectories of AQP1 diffusion on blebs generally appeared to be confined to circular areas demarcating the entire bleb surface (Fig. 2 A; see also Supplementary Material, Movie 2). AQP1 diffusion on blebs was 4–5 times faster than in the normal COS-7 plasma membrane, with D1-3 of 0.34 ± 0.05 μm2/s (143 trajectories, 13 cells). No movement of AQP1 was seen between normal plasma membrane and blebs. Because latrunculin B had no effect on AQP1 diffusion, the rapid diffusion on blebs cannot be attributed to absence of cytoskeleton. The relatively slowed AQP1 diffusion in normal plasma membrane may therefore be related to the higher density of membrane proteins than in blebs, and/or to protein/lipid organization such as rafts.
Two maneuvers were used to perturb plasma membrane lipid composition and hence lipid raft structure. The first maneuver, cyclodextrin treatment, which lowers membrane cholesterol content, reduced the average D1-3 of AQP1 by 35% in COS-7 cells (Fig. 2; Table 1; see also Supplementary Material, Movie 2). Interestingly, cyclodextrin treatment in both types of MDCK cells restricted AQP1 diffusion to a much greater extent (Fig. 2; Table 1; see also Supplementary Material, Movie 3). Following cyclodextrin treatment, >12% of AQP1 in MDCK and MDCK II cells became immobile. For the remaining mobile fraction, the average D1-3 was reduced by twofold and range by threefold.
The second maneuver, sphingomyelinase (SMase) treatment, causes the conversion of sphingomyelin (SM) to ceramide, and may also reduce cholesterol (24). SMase treatment did not affect AQP1 diffusion in COS-7 cells compared to control, untreated cells (Fig. 2, Table 1). In MDCK and MDCK II cells, SMase produced a broad distribution of diffusive behaviors, apparently further restricting some channels, while allowing faster and longer-range diffusion of a fraction AQP1. The substantial differences in AQP1 diffusion observed in COS-7 versus MDCK cells upon perturbation of lipids with CD or SMase are likely the result of differences in lipid composition of the cell plasma membranes (see Discussion).
Characterization of AQP1 diffusion by analysis of individual trajectories
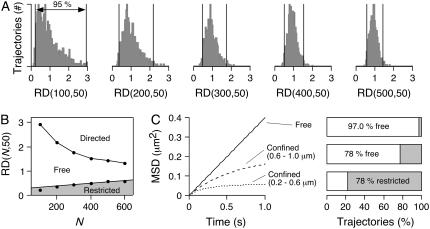
We used two independent approaches to classify modes of diffusion of individual particles, reasoning that the confidence of model-independent conclusions would be greatly strengthened. The first approach involves classification by relative deviation (RD). The RD is defined as the ratio of the experimental MSD to the line extrapolated from the initial slope of the MSD (Eq. 5), with RD < 1 indicating restricted diffusion and RD > 1 indicating directed diffusion. For nonimmobile particles, we analyzed RD at 50 frames to classify AQP1 trajectories as free, restricted, or directed. For an ensemble of particles undergoing Brownian diffusion, the distribution of RD at specified frame n broadens as total trajectory length N is reduced (29). We generated Brownian trajectories by random walk simulations, as described previously (37), using experimentally relevant trajectory lengths to establish the effective cutoff values of RD at 50 frames. Fig. 3 A shows number histograms of RD(N,50) for N in the range 100–500, with 1000 trajectories simulated for each case. The vertical black lines denote the boundaries of the central 95% of computed RD. RD values within this range were taken to represent statistical variations in Brownian motion, and those outside of the range taken as anomalous diffusion. Fig. 3 B maps the 2.5th and 97.5th percentiles of RD(N,50) as a function of N. A linear least-squares fit to the 2.5th percentile points defined the lower boundary for free diffusion, with trajectories having RD(N,50) below this line classified as restricted. Since AQP1 at the plasma membrane should not be actively directed by any cellular processes, directed diffusion was ignored, so trajectories with RD(N,50) above the 97.5th percentile were classified as free.
FIGURE 3.
Relative deviation analysis of individual simulated trajectories. (A) Histograms of trajectory number for deduced RD(N,50) for simulated free (nonanomalous) diffusion with N = 100, 200, 300, 400, and 500. Vertical lines indicate 2.5th and 97.5th percentiles. (B) RD(N,50) versus N for simulations in panel A. Solid circles indicate 2.5th and 97.5th percentiles. (C) Application of RD analysis, RD(N,50), to simulated trajectories for free diffusion that included an uncertainty of 20 nm in x- and y-coordinates. (Left) Simulations done for free diffusion (solid curve), and diffusion confined within square domains with sides of length 0.2–0.6 μm (dotted curve) or 0.6–1.0 μm (dashed curve). Simulation parameters are: 1000 trajectories, N = 300, D = 0.1 μm2/s, δt = 10 ms. (Right) Computed fractions of trajectories classified as undergoing free diffusion (white) versus restricted diffusion (gray).
To test the validity of this method of analysis, we simulated free and confined trajectories that included a 20-nm uncertainty in positional coordinates to mimic real experimental error. Positional uncertainty was simulated by distributing coordinates over a two-dimensional Gaussian profile with a standard deviation of 20 nm in x and y (38). For the simulation the diffusion coefficient was set to 0.1 μm2/s, δt was 10 ms, and N was 300. As expected, the combined MSD for simulated free diffusion was linear (Fig. 3 C). Using RD(300,50), 97.0% of the trajectories from the “free” simulation were correctly identified as free, near the 97.5% value for error-free simulations. Simulations of confined diffusion were done by restricting the trajectories to square domains with sides ranging in length from 0.2 to 0.6 μm or 0.6–1.0 μm, with 1000 trajectories generated for each group. The combined MSD of the confined trajectories shows the expected characteristic negative curvature (Fig. 3 C, left). For RD(300,50), 78% of those trajectories confined to 0.2–0.6 μm were correctly identified as restricted, whereas 78% of those for 0.6–1.0 μm were classified as free. Therefore, for particles with a diffusion coefficient of 0.1 μm2/s, the upper limit of the detectable confinement area using RD(300,50) and δt = 10 ms was ∼0.3 μm2. However, larger confinement sizes would be identifiable from longer acquisition times. For example, if δt = 30 ms under otherwise identical conditions, the upper limit of confinement identified by RD(300,50) would be ∼0.5 μm2.
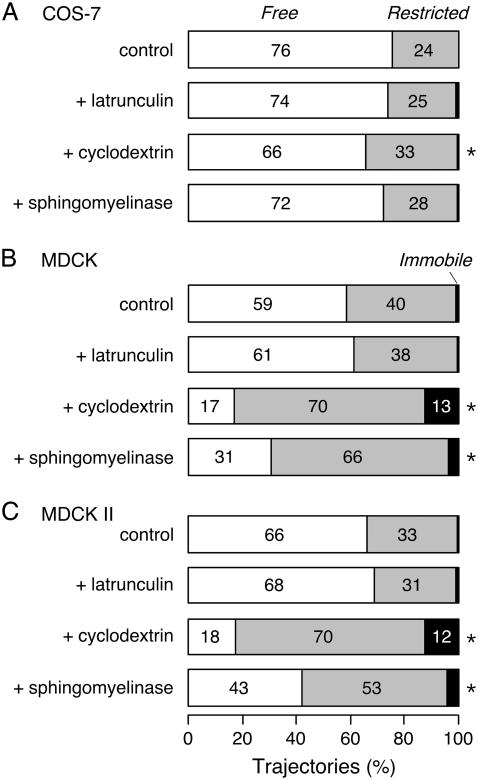
Fig. 4 summarizes the classification of experimentally measured AQP1 trajectories using RD(N,50) for data acquired at 91 Hz over 6 s and average N of 395 frames. Notably, for all three cell types under control conditions most trajectories were determined by this analysis method to be free (76% in COS-7 cells, 59% in MDCK cells, 66% in MDCK II cells). By comparison, 82% of Qdot-linked lipids were classified as free in COS-7 cells. Disruption of the cytoskeleton with latrunculin B had little effect on classifications of AQP1 diffusion. In COS-7 cells, CD and SMase treatments to alter membrane composition had little or no effect, when compared to MDCK cells, where CD and SMase produced a marked change, with >80% of trajectories becoming restricted or immobile after CD, with lesser effect of SMase. In each experiment, data were also acquired over the same image area at 32 Hz for 20 s. Classification using RD(N,50) gave an essentially identical breakdown of diffusive modes (not shown), indicating that the confinement areas (or the spacing of obstructing barriers) sampled were sufficiently small to be identified in 6 s.
FIGURE 4.
Classification of AQP1 trajectories by relative deviation analysis. RD(N,50) applied to experimental trajectories from AQP1 diffusion in COS-7 (A), MDCK (B), and MDCK II (C) cells. Percentage trajectories shown classified as free (white), restricted (gray), and immobile (black), following the indicated treatments. Immobile trajectories defined as those with range <18 nm after 100 frames. *Indicates P < 0.01 when compared to untreated cells.
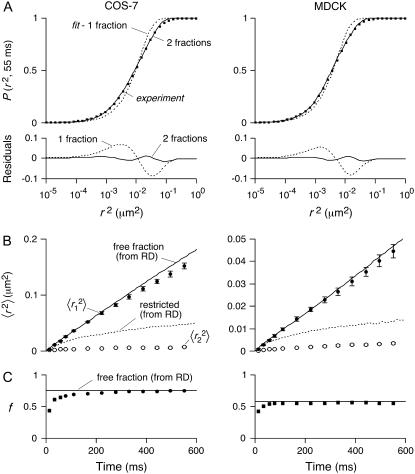
The second, independent method for analysis of individual AQP1 trajectories involved computation of cumulative probability distribution functions (CDFs) of square displacements at specified time, P(r2,t). For two populations with fractions f and (1 − f) having Brownian diffusion at different rates D1 > D2:
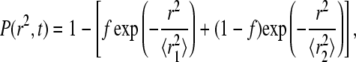 |
(6) |
where 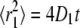 and
and 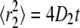 (39). Fig. 5 shows fits to CDFs for experimental AQP1 trajectories in COS-7 and MDCK cells. Fitting to Eq. 6 with two fractions provided a much better fit than to a single component. Comparison of computed
(39). Fig. 5 shows fits to CDFs for experimental AQP1 trajectories in COS-7 and MDCK cells. Fitting to Eq. 6 with two fractions provided a much better fit than to a single component. Comparison of computed 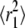 versus time shows excellent agreement with the combined MSD of trajectories that were classified as free by RD(N,50) (Fig. 5 B). There was also excellent agreement between the fitted fraction f and the fraction of trajectories classified as free by RD(N,50) at longer times, though at shorter times the correlation was poor (Fig. 5 C). The slow-diffusing constants
versus time shows excellent agreement with the combined MSD of trajectories that were classified as free by RD(N,50) (Fig. 5 B). There was also excellent agreement between the fitted fraction f and the fraction of trajectories classified as free by RD(N,50) at longer times, though at shorter times the correlation was poor (Fig. 5 C). The slow-diffusing constants 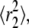 as calculated from fits to the CDF, do not agree with the combined MSD from restricted trajectories as classified by RD(N,50) (Fig. 5 B). Disagreements between these methods are due to the assumption in Eq. 6 that both components undergo Brownian diffusion, whereas in fact the second fraction of AQP1 diffused anomalously.
as calculated from fits to the CDF, do not agree with the combined MSD from restricted trajectories as classified by RD(N,50) (Fig. 5 B). Disagreements between these methods are due to the assumption in Eq. 6 that both components undergo Brownian diffusion, whereas in fact the second fraction of AQP1 diffused anomalously.
FIGURE 5.
Classification of AQP1 trajectories by analysis of cumulative distribution functions of square displacements. (A) Cumulative distributions of square displacements at 55 ms for COS-7 (left) and MDCK (right) cells. Fitting results to Eq. 6 and residuals, assuming a single Brownian fraction (f = 1, dotted curves) or two fractions (solid curves). (B) Comparison of CDF fit versus separation of individual MSDs by RD(N,50) (from Figs. 3 and 4). Fits to CDFs done at times nδt from n = 1 (11 ms) through n = 50 (550 ms). Resulting constants 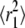 and
and 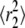 shown as solid and open circles, respectively, compared with the combined MSDs of the trajectories defined as free (solid curve) or restricted (dotted curve) by RD(N,50). (C) Fraction f of the fast component (circles) compared to the fraction of MSDs defined as free (horizontal line) by RD(N,50).
shown as solid and open circles, respectively, compared with the combined MSDs of the trajectories defined as free (solid curve) or restricted (dotted curve) by RD(N,50). (C) Fraction f of the fast component (circles) compared to the fraction of MSDs defined as free (horizontal line) by RD(N,50).
It was concluded from both methods of analysis that the majority of AQP1 diffusion in MDCK and COS-7 cells under control conditions was Brownian. Table 1 provides the results of separately analyzing the MSDs of trajectories after categorization as free or restricted by RD(N,50). In untreated cells, the mean D1-3 was ∼20% higher for AQP1 that exhibited free versus restricted diffusion (P > 0.02), whereas the mean range after 1 s of restricted AQP1 was ∼50% of that of free AQP1 (P < 0.001). No change was found in the free or restricted fractions of AQP1 upon disruption of cytoskeleton by latrunculin, but a large fraction of freely diffusing AQP1 became significantly slowed and restricted after reduction of membrane cholesterol in MDCK cells by CD.
DISCUSSION
We used Qdots to track the diffusion of AQP1 water channels in the plasma membrane of mammalian cells. The very bright fluorescence of Qdots allowed tracking at a high frame rate using wide-field microscopy at the apical membrane, where total internal reflection fluorescence is not possible. The excellent photostability of Qdots allowed the acquisition of longer trajectories than are possible with GFP or small-molecule fluorescent probes. The SPT results here confirm and extend our previous photobleaching results from GFP-AQP1 chimeras (19), which were also found to be fully mobile in the plasma membrane. The diffusion coefficient of AQP1 strongly depended on cell type, and the motion of AQP1 was largely Brownian, providing evidence against a specific association with lipid rafts or obstruction by cytoskeletal barriers.
Qdots were coupled to an engineered AQP1 mutant via antibody binding to a human c-myc epitope inserted in the second extracellular loop of each AQP1 monomer (Fig. 1 A). This site was chosen because it is well removed from any of the known functional sites in AQP1, including the glycosylation site in the first extracellular loop, the site of monomer-monomer contact, or the highly conserved NPA-NPA motif that serves as the selectivity filter. Previous functional studies showed that this epitope-tagged AQP1 forms functional water channels (21). We do not believe that attached Qdots slow membrane diffusion because of “drag” caused by the particle as it moves along the outer membrane surface. Diffusion of Qdots in solution is 2–3 orders of magnitude faster than typical membrane diffusion (40), suggesting that the membrane anchor is the limiting factor in Qdot diffusion. Further, we measured the diffusion of streptavidin-coated Qdots bound to biotinylated lipids (bi-DPPE) in COS-7 cell membranes (not shown). SPT at 91 Hz revealed that 82% of Qdot-bound bi-DPPE exhibited Brownian diffusion, with an average D1-3 of 0.21 ± 0.01 μm2/s, which is comparable to diffusion coefficients measured for cell membrane lipids covalently labeled with small fluorophores (41). Also, the substantially more rapid diffusion of AQP1-conjugated Qdots in membrane blebs compared to control plasma membrane supports the conclusion that extracellular factors do not restrict Qdot diffusion under the conditions of our experiments. Labeling here was done using a mouse anti-c-myc primary antibody, followed by F(ab′)2-linked Qdots. The particular strategy used to link Qdots did not affect diffusion because we found identical AQP1 diffusion in COS-7 cells when AQP1 was labeled with biotinylated anti-c-myc, followed by streptravidin Qdots (not shown). Antibody-linked Qdots thus provide a robust system for the study of AQP1 diffusion.
In all three cell types studied here, the majority of AQP1 exhibited Brownian diffusion. In the nonpolarized COS-7 cells, 76% of AQP1 showed unrestricted diffusion, with an average range of 0.5 μm in 1 s. Time-lapse SPT over 6 min revealed that AQP1 diffuses over the entire surface of the COS-7 cell membrane (Fig. 1 D), with individual channels moving up to 18 μm in 5 min. In MDCK and MDCK II cells, 59% and 66% of AQP1 showed free diffusion, respectively, with rates and ranges 2–3 times lower than in COS-7 cells (Table 1). The differences in the diffusion coefficient and free fraction of AQP1 in COS-7 and MDCK cells may result from differences in plasma membrane lipid composition, or perhaps the presence of microvilli on the MDCK membrane, as discussed below. To our knowledge, no other transmembrane protein has been found to exhibit this extent of unrestricted diffusion in cell membranes over such large distances. Thus, AQP1 exists in the plasma membrane largely free of specific interactions. Furthermore, our data argues against the recent suggestion of universally anomalous diffusion in cell plasma membranes (42). However, our data do not resolve whether the majority of AQP1 undergoes purely Brownian diffusion with a minor fraction undergoing anomalous diffusion, or whether the observed restricted fraction is the result of low frequency transient anomalous diffusion of many AQP1 molecules (43).
Independent methods of analysis confirmed that the majority of AQP1 undergoes Brownian diffusion in COS-7 and MDCK cells. We first used the method of Kusumi and co-workers (29), which is based on the RD of the MSD at 50 frames (Eq. 5), to classify individual trajectories as free versus restricted (Fig. 3). We also used the method of Schütz and co-workers (39), which uses a two-component fit (Eq. 6) to the CDF of square displacements (Fig. 5 A). Both analyses indicated that the majority of AQP1 diffused in a Brownian manner in COS-7 and MDCK cells, with excellent agreement in calculated MSDs by the two quite different analysis methods (Fig. 5 B). However, there was disagreement in computed MSDs for the slower or restricted fractions of AQP1 determined by RD versus CDF analysis. The assumption in the CDF model that the population is split into two uniformly Brownian fractions is the cause of this discrepancy. The second fraction probably diffuses anomalously, or may itself be composed of multiple subpopulations. A different model has been developed to describe the CDF for anomalous diffusion (44). As expected, our data also fit well to this model containing anomalous diffusion (not shown), but little useful information was obtained from the fit, other than to confirm that at least a fraction of AQP1 diffuses anomalously. If sufficient frame rate, trajectory length, and signal/noise ratio is possible, classification of individual particles by RD provides the most useful and complete information set from SPT data. An advantage of the RD method is the ability to use the individual MSDs to directly compare the rates and ranges of the diffusing fractions (Table 1).
Modulation by cytoskeleton and membrane protein content
AQP1 diffusion was not affected by disruption of the cell cytoskeleton by latrunculin B. This finding contrasts with previous SPT experiments done in our lab with CFTR, which is relatively immobile under control conditions in these same cell lines, but shows remarkable mobilization upon cytoskeletal disruption (28). Unlike AQP1, CFTR interacts with the cytoskeleton through a C-terminal PDZ-binding domain. Still, the lack of any cytoskeletal influence on AQP1 diffusion seems to contradict recent studies suggesting that the diffusion of all membrane proteins and lipids is affected by “pickets” or transmembrane “fences” that are anchored by the cytoskeleton (42). We acknowledge that our fastest acquisition rate of 11 ms is not as fast as used by Kusumi and co-workers to detect “hop diffusion”, which originally gave rise to the model of universal restriction by the cytoskeleton. However, according to this model, the depolymerization of the actin skeleton should manifest itself as a significantly higher measured diffusion coefficient for all membrane components, even at lower frame rates.
AQP1 diffusion in membrane-associated blebs was 4–5 times faster than in the normal plasma membrane of COS-7 cells (Fig. 2; see also Supplementary Material, Movie 2). Our calculated D1-3 value is likely an underestimate of the actual diffusion coefficient because the observed diffusion in blebs occurred on highly curved membrane surfaces. Blebs are cytoskeleton-free membranes that are also significantly de-enriched in membrane proteins (45,46). For example, proteins that interact directly with the cytoskeleton do not partition into blebs, which alone greatly lowers the overall membrane protein amount in blebs. The actual composition of blebs is unknown, but they do contain some proteins (other than AQP1), including LDL receptor, IgE receptor, and Con A receptor (35,45,47). Membrane blebs also contain lipid-anchored proteins such as Lyn kinase, as well as all the necessary lipids for the formation of raft-like domains (36). Taken together, our results for AQP1 diffusion in membrane blebs and latrunculin-treated cells support earlier reports that, in the absence of specific cytoskeletal binding or intermolecular interactions, diffusion is primarily controlled by the overall protein content of the plasma membrane and not by cytoskeletal barriers (46,47).
Modulation of AQP1 diffusion by cholesterol depletion
AQP1 diffusion was impeded by depletion of cholesterol with methyl-β-cyclodextrin (CD), with the extent of slowing or restriction dependent on cell type. In MDCK cells, the depletion of cholesterol by CD resulted in a greater than twofold reduction in the average AQP1 diffusion coefficient. More significantly, >87% of the AQP1 population was restricted or immobile after treatment with CD in MDCK cells, compared to only 34% in COS-7 cells (Figs. 2 and 4; Table 1; see also Supplementary Material, Movies 2 and 3). Interestingly, the observed reduction in the diffusion coefficient of AQP1 in COS-7 cells (∼35%) is nearly the same as the reduction seen in the diffusion coefficient of putative raft and nonraft proteins measured by photobleaching upon cholesterol depletion with CD in the same cell line (16).
Restricted diffusion of membrane proteins and lipids after cholesterol depletion has been observed in multiple cell systems, and is believed to result from the formation of solid-like lipid clusters in the membrane (48). The difference in the efficacy of cholesterol depletion on AQP1 diffusion in COS-7 versus MDCK cells is probably related to the lipid content of the respective plasma membranes. In particular, the ratio of glycerophospholipids (PL) to sphingomyelin (SM) and glycosphingolipids (GSL) can have a significant impact on membrane phase behavior in a cholesterol-dependent manner (49,50). The cholesterol content (30–40% of total lipid) and SM content (10–15%) of COS-7 and MDCK cell plasma membranes is nearly the same (22,51), but the GSL content of MDCK cell apical membranes is likely considerably higher than that in COS-7 cells. Measurements of budding viral particles from the apical membrane of MDCK cells indicate a GSL/PL ratio of 1:1 (52). The exact GSL content of COS-7 cells is not known, but in general, fibroblast cell lines contain much less GSL than epithelial cells. For example, GSLs compose <5% of total lipid in the plasma membrane of baby hamster kidney fibroblasts, and <1% in rat liver fibroblasts (53). Changes in cholesterol content produce major changes in membrane phase behavior, particularly if the GSL content is high. GSLs interact strongly with cholesterol and SM; at normal cholesterol levels they may form lipid rafts, or if the GSL/SM content is high enough, a continuous liquid-ordered phase. At low cholesterol, GSLs self-aggregate into gel-like phases, driven by strong H-bonding between glycosyl headgroups (52). Because GSLs likely occupy a major fraction of the membrane surface in MDCK cells, highly ordered GSL-only clusters could form a network that is nearly impenetrable to large protein complexes such as AQP1 tetramers, resulting in the observed large increase in anomalous diffusion, confinement, or immobilization. If GSLs comprise a significantly lower fraction of the total lipid in the COS-7 membrane, the solid domains that form upon cholesterol depletion will be more dispersed, resulting only in a reduction in the rate of AQP1 diffusion.
An alternate model would suggest that the restriction of membrane protein diffusion upon cholesterol depletion is the result of a reorganization or loss of the signaling lipid phosphatidylinosital-(4,5)-bisphosphate (PIP2) in the plasma membrane, which leads to a rearrangement of the actin cytoskeleton (54). If actin rearrangement upon treatment with CD limits the diffusion of AQP1, which has no specific interactions with the cytoskeleton, it must do so by clustering other membrane proteins and/or lipids, which in turn act as barriers to diffusion. According to this model, the observed differences in AQP1 diffusion upon cholesterol depletion in COS-7 and MDCK cells would indicate lower PIP2 content and/or fewer actin-binding membrane proteins in the COS-7 cell plasma membrane.
Regardless of which model is correct, AQP1 diffusion upon cholesterol depletion is likely impeded by the formation of relatively immobile barriers in the membrane. Therefore, the extreme restriction observed in the MDCK membrane can be framed in the context of excluded volume, or percolation theory (55). The percolation threshold is the concentration of barriers at which an impenetrable array is formed, completely obstructing long-range diffusion. This theory can be tested by measurement of the diffusion of species with different sizes. In a continuous fluid membrane, the size of a diffusing protein or lipid has little effect on the rate or extent of diffusion (56). However, when the membrane includes solid-like impermeable barriers, diffusion becomes strongly dependent on size (57). For a lipid, the smallest diffusing species in a membrane, the percolation threshold would be the point at which the barriers become connected. For a large tetramer such as AQP1, the effective percolation threshold is much lower. Monte-Carlo simulations indicate an approximate twofold lower percolation threshold for a diffusing particle with a twofold larger radius (57). We therefore measured the diffusion of a fluorescent lipid Rh-DPPE by photobleaching in MDCK cells before and after treatment with CD (not shown). In the normal MDCK cell membrane, the calculated diffusion coefficient of Rh-DPPE was ∼0.1 μm2/s, with a mobile fraction >90% after 1 min. We found that cholesterol depletion lowered the diffusion coefficient of the lipid probe by more than twofold, but did not significantly change the mobile fraction. Therefore, although diffusion of the lipid was impeded by cholesterol depletion, long-range diffusion was still possible, consistent with the model of volume exclusion by solid barriers. In MDCK cells, the fraction of the membrane occupied by solid-like domains formed upon cholesterol depletion must lie above the effective percolation threshold for AQP1, but below the actual percolation threshold sensed by lipids.
Modulation of diffusion by sphingomyelin hydrolysis
Sphingomyelinase (SMase) converts SM to ceramide by hydrolysis of the headgroup at the phosphate-ester bond. In MDCK cells, SMase treatment produced a broad distribution of AQP1 diffusive behaviors (Fig. 2, B and C). An increase in the percentage of restricted trajectories (Fig. 4, B and C) was accompanied by the appearance of a subpopulation of AQP1 that exhibited diffusion 4–5 times faster than in the normal membrane. In COS-7 cells, SMase had no measurable effect on AQP1 diffusion.
Like GSLs, ceramide segregates from SM/cholesterol lipid rafts to form solid-like domains in membranes (58). SMase treatment may also reduce membrane cholesterol (24). Therefore, the appearance of a diffusion-restricted fraction of AQP1 after SMase treatment is likely the result of interference from solid-like domains of ceramide and/or GSL. The appearance of a faster diffusing fraction of AQP1 is probably the result of lipid “scrambling” across the membrane upon treatment with SMase (59). A loss of lipid asymmetry in the plasma membrane could result in the disruption of protein-protein or protein-lipid interactions on a large scale, leading to significantly protein de-enriched membrane domains. It could also lead to a disruption of ordered lipid phases found in the outer leaflet of the plasma membrane, leading to a less viscous membrane. Disruption of lipid raft-associated components may also lead to the reduction of microvilli in the MDCK membrane (60). These events alone, or in combination, could account for the de novo appearance of the subpopulation of AQP1 with relatively fast diffusion.
CONCLUSIONS
We examined the diffusion of AQP1 in the plasma membranes of well-characterized cell lines under various conditions to look for evidence of confinement by cholesterol-rich lipid rafts, the cytoskeleton, or other membrane proteins. We found that under control conditions most AQP1 diffused freely, covering up to several microns per minute. This behavior contrasts with previously reported SPT experiments on putative raft-associated proteins and lipids, which are generally confined in cholesterol-dependent domains, resulting in mostly anomalous diffusion (12,15). Our data therefore argue against the accumulation of AQP1 in lipid rafts in the cell membrane, despite its recovery in the detergent-insoluble fractions of cells. Depolymerization of the actin cytoskeleton had no effect on the diffusion of AQP1, but diffusion could be altered with other treatments. The rate of AQP1 diffusion was greatly increased in membrane blebs where overall protein content is lower than in the normal plasma membrane, whereas it was obstructed by solid-like lipid or protein clusters created by cholesterol depletion. However, these effects are nonspecific, as they have been observed universally for raft and nonraft associated proteins, signaling complexes, and multiple receptors (16,24,25,35,45,47,54). We conclude that AQP1 is a largely noninteracting protein, whose diffusion is mostly Brownian and determined by the concentration of obstructions in the plane of the membrane.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Dr. Songwan Jin for his contribution of simulated trajectories. We thank Dr. Peter Haggie for his contribution of DNA constructs and useful comments.
This work was supported by National Institutes of Health grants EB00415, HL73856, DK35124, HL59198, and EY13574, Cystic Fibrosis Research and Translational Core Center grant DK72517, and Research Development Program and Drug Discovery grants from the Cystic Fibrosis Foundation. Dr. Crane was supported in part by National Institutes of Health National Research Service Award GM808512.
Editor: Thomas Schmidt.
References
- 1.Verkman, A. S. 2005. More than just water channels: unexpected cellular roles of aquaporins. J. Cell Sci. 118:3225–3232. [DOI] [PubMed] [Google Scholar]
- 2.Verbavatz, J. M., D. Brown, I. Sabolic, G. Valenti, D. A. Ausiello, A. N. Van Hoek, T. Ma, and A. S. Verkman. 1993. Tetrameric assembly of CHIP28 water channels in liposomes and cell membranes: a freeze-fracture study. J. Cell Biol. 123:605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren, G., A. Cheng, P. Melnyk, and A. K. Mitra. 2000. Polymorphism in the packing of aquaporin-1 tetramers in 2-D crystals. J. Struct. Biol. 130:45–53. [DOI] [PubMed] [Google Scholar]
- 4.Sui, H., B. G. Han, J. K. Lee, P. Walian, and B. K. Jap. 2001. Structural basis of water-specific transport through the AQP1 water channel. Nature. 414:872–878. [DOI] [PubMed] [Google Scholar]
- 5.Sabolic, I., G. Valenti, J. M. Verbavatz, A. N. Van Hoek, A. S. Verkman, D. A. Ausiello, and D. Brown. 1992. Localization of the CHIP28 water channel in rat kidney. Am. J. Physiol. 263:C1225–C1233. [DOI] [PubMed] [Google Scholar]
- 6.Zhang, R., W. Skach, H. Hasegawa, A. N. van Hoek, and A. S. Verkman. 1993. Cloning, functional analysis and cell localization of a kidney proximal tubule water transporter homologous to CHIP28. J. Cell Biol. 120:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen, S., T. Pallone, B. L. Smith, E. I. Christensen, P. Agre, and A. B. Maunsbach. 1995. Aquaporin-1 water channels in short and long loop descending thin limbs and in descending vasa recta in rat kidney. Am. J. Physiol. 268:F1023–F1037. [DOI] [PubMed] [Google Scholar]
- 8.Speake, T., L. J. Freeman, and P. D. Brown. 2003. Expression of aquaporin 1 and aquaporin 4 water channels in rat choroid plexus. Biochim. Biophys. Acta. 1609:80–86. [DOI] [PubMed] [Google Scholar]
- 9.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–572. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi, H., H. Yokoo, T. Yanagita, S. Satoh, B. Kis, M. Deli, M. Niwa, and A. Wada. 2006. Induction of aquaporin 1 by dexamethasone in lipid rafts in immortalized brain microvascular endothelial cells. Brain Res. 1123:12–19. [DOI] [PubMed] [Google Scholar]
- 11.Palestini, P., C. Calvi, E. Conforti, R. Daffara, L. Botto, and G. Miserocchi. 2003. Compositional changes in lipid microdomains of air-blood barrier plasma membranes in pulmonary interstitial edema. J. Appl. Physiol. 95:1446–1452. [DOI] [PubMed] [Google Scholar]
- 12.Sheets, E. D., G. M. Lee, R. Simson, and K. Jacobson. 1997. Transient confinement of a glycosylphosphatidylinositol-anchored protein in the plasma membrane. Biochemistry. 36:12449–12458. [DOI] [PubMed] [Google Scholar]
- 13.Schutz, G. J., G. Kada, V. P. Pastushenko, and H. Schindler. 2000. Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J. 19:892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vrljic, M., S. Y. Nishimura, S. Brasselet, W. E. Moerner, and H. M. McConnell. 2002. Translational diffusion of individual class II MHC membrane proteins in cells. Biophys. J. 83:2681–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich, C., B. Yang, T. Fujiwara, A. Kusumi, and K. Jacobson. 2002. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 82:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenworthy, A. K., B. J. Nichols, C. L. Remmert, G. M. Hendrix, M. Kumar, J. Zimmerberg, and J. Lippincott-Schwartz. 2004. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 165:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meder, D., M. J. Moreno, P. Verkade, W. L. Vaz, and K. Simons. 2006. Phase coexistence and connectivity in the apical membrane of polarized epithelial cells. Proc. Natl. Acad. Sci. USA. 103:329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho, M. R., D. W. Knowles, B. L. Smith, J. J. Moulds, P. Agre, N. Mohandas, and D. E. Golan. 1999. Membrane dynamics of the water transport protein aquaporin-1 in intact human red cells. Biophys. J. 76:1136–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umenishi, F., J. M. Verbavatz, and A. S. Verkman. 2000. cAMP regulated membrane diffusion of a green fluorescent protein-aquaporin 2 chimera. Biophys. J. 78:1024–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin, M. H., P. M. Haggie, L. Vetrivel, and A. S. Verkman. 2001. Diffusion in the endoplasmic reticulum of an aquaporin-2 mutant causing human nephrogenic diabetes insipidus. J. Biol. Chem. 276:21331–21336. [DOI] [PubMed] [Google Scholar]
- 21.Lu, Y., I. R. Turnbull, A. Bragin, K. Carveth, A. S. Verkman, and W. R. Skach. 2000. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol. Biol. Cell. 11:2973–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 68:533–544. [DOI] [PubMed] [Google Scholar]
- 23.Deen, P. M., S. Nielsen, R. J. Bindels, and C. H. van Os. 1997. Apical and basolateral expression of aquaporin-1 in transfected MDCK and LLC-PK cells and functional evaluation of their transcellular osmotic water permeabilities. Pflugers Arch. 433:780–787. [DOI] [PubMed] [Google Scholar]
- 24.Vrljic, M., S. Y. Nishimura, W. E. Moerner, and H. M. McConnell. 2004. Cholesterol depletion suppresses the translational diffusion of class II major histocompatibility complex proteins in the plasma membrane. Biophys. J. 88:334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodwin, J. S., K. R. Drake, C. L. Remmert, and A. K. Kenworthy. 2005. Ras diffusion is sensitive to plasma membrane viscosity. Biophys. J. 89:1398–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiwara, T., K. Ritchie, H. Murakoshi, K. Jacobson, and A. Kusumi. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157:1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxton, M. J. 1997. Single-particle tracking: the distribution of diffusion coefficients. Biophys. J. 72:1744–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haggie, P. M., J. K. Kim, G. L. Lukacs, and A. S. Verkman. 2006. Tracking of quantum dot-labeled CFTR shows near immobilization by C-terminal PDZ interactions. Mol. Biol. Cell. 17:4937–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusumi, A., Y. Sako, and M. Yamamoto. 1993. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65:2021–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Axelrod, D., D. E. Koppel, J. Schlessinger, E. Elson, and W. W. Webb. 1976. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16:1055–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yguerabide, J., J. A. Schmidt, and E. E. Yguerabide. 1982. Lateral mobility in membranes as detected by fluorescence recovery after photobleaching. Biophys. J. 40:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sako, Y., A. Nagafuchi, S. Tsukita, M. Takeichi, and A. Kusumi. 1998. Cytoplasmic regulation of the movement of E-cadherin on the free cell surface as studied by optical tweezers and single particle tracking: corralling and tethering by the membrane skeleton. J. Cell Biol. 140:1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choquet, D., and A. Triller. 2003. The role of receptor diffusion in the organization of the postsynaptic membrane. Nat. Rev. Neurosci. 4:251–265. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, K., K. Ritchie, E. Kajikawa, T. Fujiwara, and A. Kusumi. 2005. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 88:3659–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tank, D. W., E. S. Wu, and W. W. Webb. 1982. Enhanced molecular diffusibility in muscle membrane blebs: release of lateral constraints. J. Cell Biol. 92:207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumgart, T., A. T. Hammond, P. Sengupta, S. T. Hess, D. A. Holowka, B. A. Baird, and W. W. Webb. 2007. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. USA. 104:3165–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin, S., and A. S. Verkman. 2007. Single particle tracking of complex diffusion in membranes: simulation and detection of barrier, raft, and interaction phenomena. J. Phys. Chem. B. 111:3625–3632. [DOI] [PubMed] [Google Scholar]
- 38.Jin, S., P. M. Haggie, and A. S. Verkman. 2007. Single-particle tracking of membrane protein diffusion in a potential: simulation, detection, and application to confined diffusion of CFTR Cl− channels. Biophys. J. 93:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schutz, G. J., H. Schindler, and T. Schmidt. 1997. Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys. J. 73:1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swift, J. L., R. Heuff, and D. T. Cramb. 2006. A two-photon excitation fluorescence cross-correlation assay for a model ligand-receptor binding system using quantum dots. Biophys. J. 90:1396–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murase, K., T. Fujiwara, Y. Umemura, K. Suzuki, R. Iino, H. Yamashita, M. Saito, H. Murakoshi, K. Ritchie, and A. Kusumi. 2004. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys. J. 86:4075–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kusumi, A., C. Nakada, K. Ritchie, K. Murase, K. Suzuki, H. Murakoshi, R. S. Kasai, J. Kondo, and T. Fujiwara. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. [DOI] [PubMed] [Google Scholar]
- 43.Saxton, M. J. 2007. A biological interpretation of transient anomalous subdiffusion. I. Qualitative model. Biophys. J. 92:1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deverall, M. A., E. Gindl, E. K. Sinner, H. Besir, J. Ruehe, M. J. Saxton, and C. A. Naumann. 2005. Membrane lateral mobility obstructed by polymer-tethered lipids studied at the single molecule level. Biophys. J. 88:1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barak, L. S., and W. W. Webb. 1982. Diffusion of low density lipoprotein-receptor complex on human fibroblasts. J. Cell Biol. 95:846–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frick, M., K. Schmidt, and B. J. Nichols. 2007. Modulation of lateral diffusion in the plasma membrane by protein density. Curr. Biol. 17:462–467. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, J. L., T. J. Feder, and W. W. Webb. 1992. Effects of protein concentration on IgE receptor mobility in rat basophilic leukemia cell plasma membranes. Biol. Cell. 61:1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishimura, S. Y., M. Vrljic, L. O. Klein, H. M. McConnell, and W. E. Moerner. 2005. Cholesterol depletion induces solid-like regions in the plasma membrane. Biophys. J. 90:927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sankaram, M. B., and T. E. Thompson. 1990. Interaction of cholesterol with various glycerophospholipids and sphingomyelin. Biochemistry. 29:10670–10675. [DOI] [PubMed] [Google Scholar]
- 50.Silvius, J. R. 1992. Cholesterol modulation of lipid intermixing in phospholipid and glycosphingolipid mixtures. Evaluation using fluorescent lipid probes and brominated lipid quenchers. Biochemistry. 31:3398–3408. [DOI] [PubMed] [Google Scholar]
- 51.Matthews, V., B. Schuster, S. Schutze, I. Bussmeyer, A. Ludwig, C. Hundhausen, T. Sadowski, P. Saftig, D. Hartmann, K. J. Kallen, and S. Rose-John. 2003. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE). J. Biol. Chem. 278:38829–38839. [DOI] [PubMed] [Google Scholar]
- 52.Simons, K., and G. van Meer. 1988. Lipid sorting in epithelial cells. Biochemistry. 27:6197–6202. [DOI] [PubMed] [Google Scholar]
- 53.Renkonen, O., C. G. Gahmberg, K. Simons, and L. Kaariainen. 1972. The lipids of the plasma membranes and endoplasmic reticulum from cultured baby hamster kidney cells (BHK21). Biochim. Biophys. Acta. 255:66–78. [DOI] [PubMed] [Google Scholar]
- 54.Kwik, J., S. Boyle, D. Fooksman, L. Margolis, M. P. Sheetz, and M. Edidin. 2003. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA. 100:13964–13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saxton, M. J. 1994. Anomalous diffusion due to obstacles: a Monte Carlo study. Biophys. J. 66:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saffman, P. G., and M. Delbruck. 1975. Brownian motion in biological membranes. Proc. Natl. Acad. Sci. USA. 72:3111–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saxton, M. J. 1993. Lateral diffusion in an archipelago. Dependence on tracer size. Biophys. J. 64:1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva, L. C., R. F. de Almeida, B. M. Castro, A. Fedorov, and M. Prieto. 2007. Ceramide-domain formation and collapse in lipid rafts: membrane reorganization by an apoptotic lipid. Biophys. J. 92:502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Contreras, F. X., A. V. Villar, A. Alonso, R. N. Kolesnick, and F. M. Goni. 2003. Sphingomyelinase activity causes transbilayer lipid translocation in model and cell membranes. J. Biol. Chem. 278:37169–37174. [DOI] [PubMed] [Google Scholar]
- 60.Poole, K., D. Meder, K. Simons, and D. Muller. 2004. The effect of raft lipid depletion on microvilli formation in MDCK cells, visualized by atomic force microscopy. FEBS Lett. 565:53–58. [DOI] [PubMed] [Google Scholar]