Abstract
Epstein–Barr virus (EBV) infection is associated with several human diseases that involve unrestricted proliferation of B lymphocytes. EBV nuclear antigen 1 (EBNA-1) is expressed in all EBV-infected cells and plays an essential role in persistence of the EBV genome. EBNA-1 has also been reported to have oncogenic potential. As an approach for treating EBV infections, we examined the capacity of EBNA-1 ribozymes delivered by recombinant adenoviruses to suppress EBNA-1 expression and to block virus-induced B cell proliferation. In contrast to primary B cells, EBV-transformed B lymphoblastoid cell lines expressed αv integrins, the adenovirus internalization receptors, and were also susceptible to adenovirus-mediated gene delivery. Adenovirus delivery of a specific ribozyme (RZ1) to lymphoblastoid cell lines, suppressed EBNA-1 mRNA and protein expression, significantly reduced the number of EBV genomes, and nearly abolished cell proliferation in low serum. Adenovirus delivery of RZ1 also prevented EBV infection of an established EBV-negative B cell line. These studies demonstrate the potential use of adenovirus-encoded ribozymes to treat EBV-induced lymphoproliferative disorders.
Epstein–Barr virus (EBV), a member of the herpesvirus family, persists in latently infected B cells for the life of the host (1). EBV infection occurs early in life and can cause infectious mononucleosis, a self-limiting lymphoproliferative disorder (2). EBV infection is not usually associated with significant morbidity. However, under certain conditions, infection is associated with the development of several human malignancies, including endemic Burkitt lymphoma, nasopharyngeal carcinoma, AIDS-associated lymphomas, and lymphoproliferative disorders in transplant recipients (3). A common feature associated with EBV-induced lymphoproliferative diseases is immunosuppression of the host (4, 5). EBV-specific cytotoxic T cells prevent outgrowth of EBV-infected B cells in vivo (6–8) and the absence or impairment of the immune system contributes to the development of lymphoproliferative disease. In vitro, EBV infects human peripheral blood B cells and, under appropriate conditions, immortalizes these cells termed lymphoblastoid cell lines (LCL). The proliferative potential of EBV-infected cells and lifetime exposure through persistence are major predisposing factors for the development of EBV-associated neoplasia.
Latently infected B cells contain multiple copies of the viral genome that primarily exist as episomes that replicate synchronously with cell division (9). While the EBV genome encodes ≈100 genes, viral gene expression in latently infected B cells is restricted to eight viral genes including six nuclear antigens [EBV nuclear antigen 1 (EBNA-1), -2, -3A, -3B, -3C, and -LP], two latent membrane proteins (LMP-1 and LMP-2), and two untranslated RNAs (EBER1 and EBER2) (9). EBNA-2 and LMP-1 have a direct role in cell immortalization (10, 11) and recent studies have also suggested that expression of the EBNA-1 protein alone may also contribute to B cell neoplasia (12, 13).
The EBNA-1 protein localizes to the nucleus of all cells carrying the viral genome and is required for replication and maintenance of the EBV genome (14, 15). EBNA-1 interacts with multiple binding sites in the latent cycle origin of EBV replication (oriP) and also transactivates the enhancers of a number of other viral genes (14, 16, 17).
In the studies reported here, we sought to determine whether specific ribozymes could be used to inhibit EBNA-1 expression, thereby preventing EBV-induced cell proliferation. To accomplish this goal, we examined the capacity of adenoviral vectors to deliver functional EBNA-1 specific ribozymes to LCL. Our studies indicate that EBV-transformed lymphocytes up-regulate αv integrin expression, thus increasing the efficiency with which adenovirus-delivered ribozymes inhibit EBV-induced cell proliferation.
MATERIALS AND METHODS
Cell Lines, EBV Infection, and Flow Cytometry.
The human myeloma cell line RPMI 8226 and the Burkitt lymphoma cell line Ramos (American Type Tissue Culture Collection), were cultured in RPMI 1640 medium containing 10% fetal calf serum (FCS). Peripheral blood B cells were isolated from adult donors by sedimentation on Ficoll/Hypaque density gradients (d = 1.077) followed by negative selection of monocytes and T cells as described (18). B95–8 culture supernatants containing the transforming strain of EBV were used to establish LCL (19). LCL express type 3 latency genes (EBNA-2 and LMP-1) and were maintained in RPMI medium 1640 with 5% FCS. Expression of αv integrins on B cells was analyzed by flow cytometry. Cells were incubated with 10 μg/ml of anti-αvβ3 (LM609) or anti-αvβ5 (P1F6) monoclonal antibody (kindly provided by David Cheresh, The Scripps Research Institute) or PBS alone (control) followed by incubation with fluorescein-conjugated anti-mouse IgG (Kirkegaard & Perry Laboratories). Cell samples were then analyzed by flow cytometry (FACScan II, Becton Dickinson). In separate studies, freshly isolated peripheral blood lymphocytes or tonsil lymphocytes were sorted into separate cell populations that expressed or lacked αv integrins using a polyclonal anti-αv antibody (Life Technologies, Grand Island, NY). Each cell population was then analyzed for the presence of EBV by immunoblotting as described below.
Construction of EBNA-1 Ribozymes and in Vitro RNA Cleavage Assay.
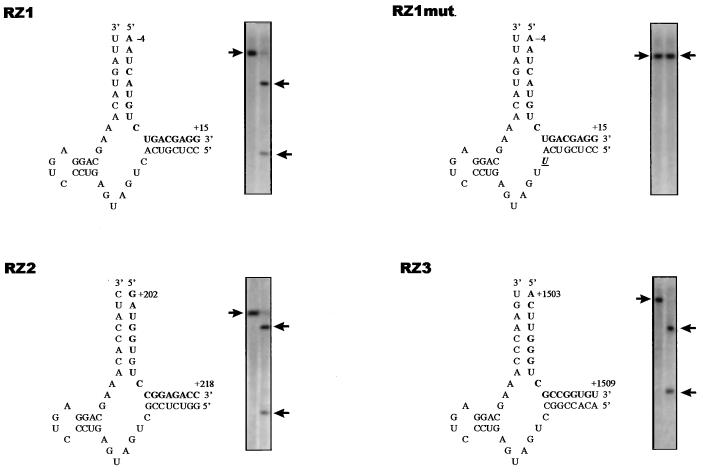
Three hammerhead ribozymes, designated RZ1, RZ2, and RZ3 (Fig. 1), were designed (20) to hydrolyze the GUC sequence at nucleotide positions 3–6, 308–310, and 1509–1511 relative to the translation initiation site in EBNA-1 RNA, respectively. A mutant ribozyme, RZ1 mut, was constructed by introducing a single base mutation in the catalytic site of RZ1 at position 9 (C to U). Synthetic DNA oligonucleotides encoding each of the EBNA-1 ribozymes, as well as their complementary strands (Applied Biosystems), were hybridized and then cloned into the HindIII–KpnI (RZ1), BamHI–EcoRI (RZ2), and EcoRV–NotI (RZ3) sites in pcDNA3 (Invitrogen). Each of the plasmid constructs were sequenced to confirm the correct orientation and DNA sequence of the ribozymes.
Figure 1.
Structure and functional activity of EBNA-1 hammerhead ribozymes. The GUC cleavage sites recognized by ribozymes RZ1, RZ1 mut, RZ2, and RZ3 in EBNA-1 RNA (boldface), and the in vitro RNA cleavage activity of each ribozyme is shown. The arrows on the left indicate the location of template RNA incubated in the absence of ribozymes while the arrows on the right indicate the migration of RNA cleavage products generated by incubation of the RNA templates with ribozymes.
Two separate EBNA-1 cDNA fragments containing nucleotides 5518–6185 (pEB1) and 6978–7651 (pEB2) in pCEP4 (Invitrogen) were cloned into pBluescript (Stratagene) and subsequently used as substrates for in vitro RNA cleavage assays. pEB1 contains the cleavage sites for RZ1 and RZ2 while pEB2 contains the cleavage site for RZ3. EBNA-1 ribozymes were expressed from the T7 promoter in pcDNA3 by in vitro transcription (Stratagene, SD). Radiolabeled EBNA-1 transcripts were similarly synthesized by incorporating 32P-UTP. EBNA-1 ribozymes and labeled EBNA-1 transcripts were incubated at a ratio of 10:1 in the presence of 10 mM MgCl2 and 50 mM Tris⋅HCl, pH 7.4, for 1 hr at 37°C, followed by the addition of 20 mM EDTA to terminate the reaction. Reaction mixtures were electrophoresed on 6% polyacrylamide sequencing gels. RNA cleavage was quantitated by scanning dried gels on a PhosphorImager (Molecular Dynamics).
Construction of Recombinant Adenovirus Vectors.
The adenovirus shuttle vector pAd/RSV, was generated by subcloning the Rous sarcoma virus (RSV) promoter/BGH Poly(A) expression cassette from pRC/RSV (Invitrogen) into pΔE1sp1B (Microbix, Toronto). A cDNA-encoding green fluorescence protein (GFP) (pEGFP, CLONTECH) was cloned into pAd/RSV. A recombinant adenovirus containing GFP was then generated by cotransfecting 293 cells with 15 μg of pAd/RSV.GFP and an equivalent amount of the adenovirus-packaging plasmid pJM17 (Microbix) using calcium phosphate. Recombinant adenoviruses were generated by homologous recombination and isolated by plaque formation on 293 cells as described (21). GFP-producing viruses were identified by fluorescence microscopy and a high-titered virus stock of one of these, designated Ad.GFP, was propagated in 293 cells (21).
The EBNA-1 ribozyme sequences derived from the pcDNA3 constructs were cloned into pAd/RSV. Recombinant adenoviruses were then generated by cotransfection with pJM17 as described above. Insertion of the RZ sequences was determined by endonuclease restriction digestion and DNA sequencing.
RT-PCR, Southern Blot Analysis, and Immunoblotting.
Total RNA was isolated from LCL cells (Trizol reagent, GIBCO/BRL) 48 hr postinfection with 500 plaque-forming units per cell of Ad.RZ or with a control virus, Ad.RSV. Two micrograms of total RNA from each sample was then treated with RNase-free DNAase 1 (Promega), extracted with phenol:chloroform and then used as a template for cDNA synthesis using Moloney murine leukemia virus reverse transcriptase (GIBCO/BRL). The reaction mixture was heated to 95°C for 5 min and then subjected to 30–40 cycles of PCR (94°C, 1 min; 55°C, 1 min; 72°C, 1 min). To analyze expression of EBNA-1 ribozymes, PCR reactions were performed using Adv.for (5′-TTTGACCATTCACCACATTGG-3′) and Adv.rev (5′-AGATGGCTGGCAACTAGAAG-3′) oligonucleotide primers that correspond to nucleotides 574–594 and 731–750 of the pRc/RSV vector, respectively. To analyze EBNA-1 mRNA expression, PCR reactions were performed using primers EBa (5′-GGTAGAGGACGTGAAAGAGCCAG-3′) and EBb (5′-ATCGCATCCTTCAAAACCTCAGC-3′) corresponding to nucleotides 109,012–109,685 in B95–8 EBV. Expression of glyceraldehyde-3-phosphate dehydrogenase mRNA was analyzed using PCR primers Ga (5′-CTGAGAACGGGAAGCTTGTC-3′) and Gb (5′-CCTGCTTCACCACCTTCTTG-3′). Reaction products were analyzed on 1.2% agarose gels that were stained with ethidium bromide.
To analyze persistence of the EBV genome in LCL cells, genomic DNA was isolated (Micro-Turbogen, Invitrogen) from cell cultures at weekly intervals following infection with Ad.RSV.RZ or with Ad.RSV. During this time, cells were maintained in media containing 5% FCS, a condition that allowed normal growth of ribozyme-treated LCL. Twenty micrograms of BamHI digested DNA was electrophoresed on a 1% agarose gel, transferred to a nylon membrane (Nytran, Schleicher & Schuell), and then probed with an EBV BamHI W fragment.
To study the effect of EBNA-1 ribozymes on EBNA-1 protein expression, LCL cells were infected at an MOI of 500 plaque-forming units per cell with Ad.RZ1, Ad.RZ1 mut or Ad.RSV medium containing in 5% FCS. At varying times after infection, 1 × 106 cells were solubilized in lysis buffer (50 mM Tris⋅HCl, pH 7.4, containing 0.1% SDS/0.5% sodium deoxycholate/1.0% Nonidet P-40/100 μg/ml phenylmethylsulfonyl fluoride/1 μg/ml aprotinin/0.02% NaN3). Fifty micrograms of each cell lysate was electrophoresed on SDS/10% polyacrylamide gel and then transferred to a polyvinylidene difluoride membrane (Immobilon P, Millipore). The membrane was then incubated with a 1:200 dilution of an EBV-positive human serum capable or recognizing EBNA-1 (kindly provided by Lawrence Young, University of Birmingham, Birmingham, U.K.). The immunoblot was developed using an enhanced chemiluminescence detection system (Amersham).
EBNA-1 ribozymes were also analyzed for their ability to prevent EBV infection of two EBV-negative cell lines, Ramos and RPMI 8226. Each of these cell types were infected with Ad.RZ1 or a control viral vector, Ad.RSV, for 24 hr prior to the addition of B95–8 EBV supernatants. At various times after EBV infection, genomic DNA was isolated and the presence of the EBV genome was analyzed by PCR using EBNA-1 primers, EBa and EBb.
Adenovirus Gene Delivery and Cell Proliferation Assays.
The susceptibility of B cells to adenovirus-mediated gene delivery was determined by infecting cells with 500 plaque-forming units per cell of Ad.GFP for 4 hr. Unbound virus was removed by washing the cells and expression of GFP was then analyzed 36 hr postinfection by flow cytometry. The effect of EBNA-1 ribozymes on cell proliferation was analyzed as follows. LCL cells were infected with 500 plaque-forming units per cell of Ad.RZ1, Ad.RZ1 mut, or Ad.RSV for 14 days in RPMI medium 1640 containing 5% FCS. The cells were then washed and recultured in media containing 1% FCS (low serum). DNA synthesis was then quantitated at varying times by measuring BrdUTP incorporation in an ELISA (Boehringer Mannheim). In parallel studies, the total number of cells in each culture was determined by counting using light microscopy. Cell viability was ascertained by trypan blue dye exclusion.
RESULTS
RNA Cleavage Activity of EBNA-1 Ribozymes.
Hammerhead ribozymes cleave RNA targets containing the consensus sequence NUX (X = A, C, or U). Based on sequence analysis and RNA folding predictions (20), we chose three sites in EBNA-1 RNA as potential targets for ribozyme interaction (Fig. 1). Synthetic ribozymes were assayed for their ability to cleave EBNA-1 RNA in vitro. As shown in Fig. 1, RZ1, RZ2, and RZ3 were capable of cleaving EBNA-1 transcripts containing the appropriate target sequence. More than 90% of the EBNA-1 transcripts were cleaved yielding the expected size for RNA cleavage fragments. In contrast, a single base substitution introduced into the catalytic site of RZ1 (RZ1 mut) abrogated EBNA-1 RNA cleavage (Fig. 1).
αv Integrin Expression Facilitates Adenovirus-Mediated Gene Delivery to EBV-Transformed B Cells.
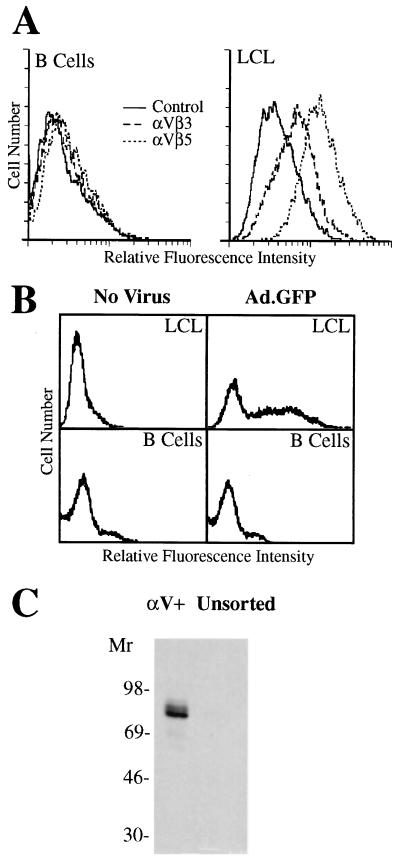
Initial studies examined whether recombinant adenovirus could be used to deliver ribozymes into transformed B cells. Adenovirus has been used to efficiently deliver antiviral ribozymes to human liver cells (22); however, previous studies indicated that peripheral blood monocytes and T lymphocytes express only very low levels of αv integrins (23), the adenovirus internalization receptor (24), and that these cells are relatively resistant to adenovirus-mediated gene delivery. We examined whether primary B lymphocytes and LCL expressed αv integrins and also whether they were susceptible to adenovirus-mediated gene delivery. Primary B lymphocytes were found to express very low levels of αv integrins (Fig. 2A) and were resistant to adenovirus-mediated gene delivery (Fig. 2B). In contrast, αv integrins were up-regulated on in vitro EBV-transformed B cells (LCL) (Fig. 2A); and these cells were susceptible to adenovirus-mediated delivery (Fig. 2B). Approximately 50% of LCL cells expressed recombinant GFP 36 hr postinfection (Fig. 2B) and ≈100% of the cells expressed GFP 7 days after infection (data not shown). Similar results were obtained with EBV-transformed B cells from five different human donors.
Figure 2.
Integrin αv expression promotes adenovirus-mediated gene delivery to human B lymphocytes. Primary human B cells or LCL cells were analyzed for αvβ3 or αvβ5 integrin expression (A) and for susceptibility to adenovirus-mediated gene delivery as examined by flow cytometry (B). The presence of EBV in freshly isolated (unsorted) lymphocytes or in αv+ sorted cells was determined by immunoblotting using a polyclonal anti-EBNA-1 antiserum (C).
We also examined whether αv integrin expression is similarly up-regulated on B cells transformed with EBV in vivo. Peripheral blood mononuclear cells were selected for αv integrin expression by flow cytometry and the presence of EBV in the αv-expressing (αv+) and the unsorted population was then analyzed. EBV was detected in αv integrin-expressing cells but not in the unsorted lymphocytes as assessed by immunoblot detection of the EBNA-1 protein (Fig. 2C). Together, these studies indicate that EBV infection is associated with up-regulation of αv integrins on B cells and this correlates with susceptibility to adenovirus-mediated gene delivery.
Expression of the EBNA-1 Ribozyme RZ1 Protects B Cells from EBV Infection.
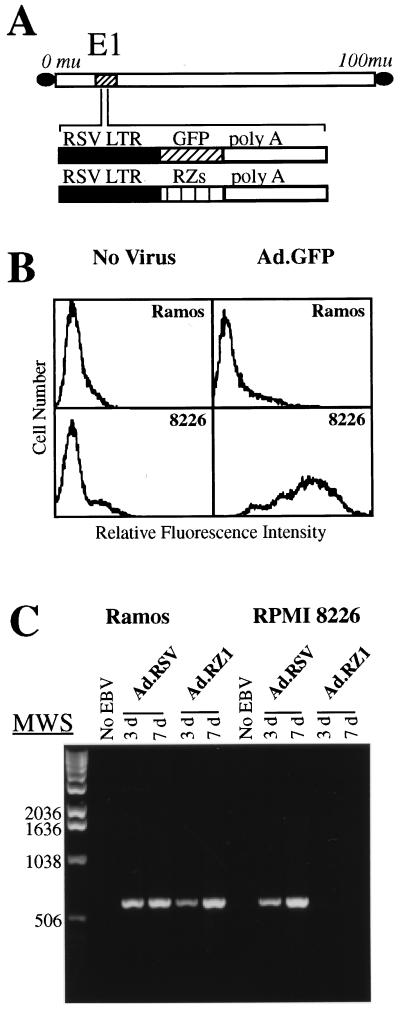
Studies were next performed to determine whether adenovirus-delivery of EBNA-1 ribozymes could prevent subsequent EBV infection of two EBV-negative B cell lines, Ramos and RPMI 8226. Both cell types express the EBV receptor, CD21 (CR2), and each can be infected by EBV; however only RPMI 8226 cells express αv integrins (25). Only RPMI 8226 cells supported adenovirus-mediated gene delivery (Fig. 3B). Furthermore, delivery of the RZ1 EBNA-1 ribozyme into RPMI 8226 cells prevented subsequent infection by EBV as determined by PCR amplification of the EBV genome. In contrast, αv integrin-negative Ramos cells were not protected from EBV infection by adenovirus delivery of RZ1 (Fig. 3C).
Figure 3.
Protection of B lymphoblastoid cell lines from EBV infection with an adenovirus-delivered EBNA-1 ribozyme. (A) Schematic representation of recombinant adenoviruses encoding GFP or EBNA-1 ribozymes (RZ). (B) Ramos (αv integrin negative) or RPMI 8226 cells (αv integrin-positive) B cells were analyzed for their susceptibility to adenovirus gene delivery by flow cytometry. (C) Ramos or RPMI 8226 cells were infected with Ad.RZ1 or a control virus (Ad.RSV) and then incubated with EBV or without (no EBV). The presence of the EBNA-1 gene, indicative of EBV infection, was analyzed 3 or 7 days later by PCR.
Effect of EBNA-1 Ribozymes on EBNA-1 Expression and Persistence of the EBV Genome in LCL Cells.
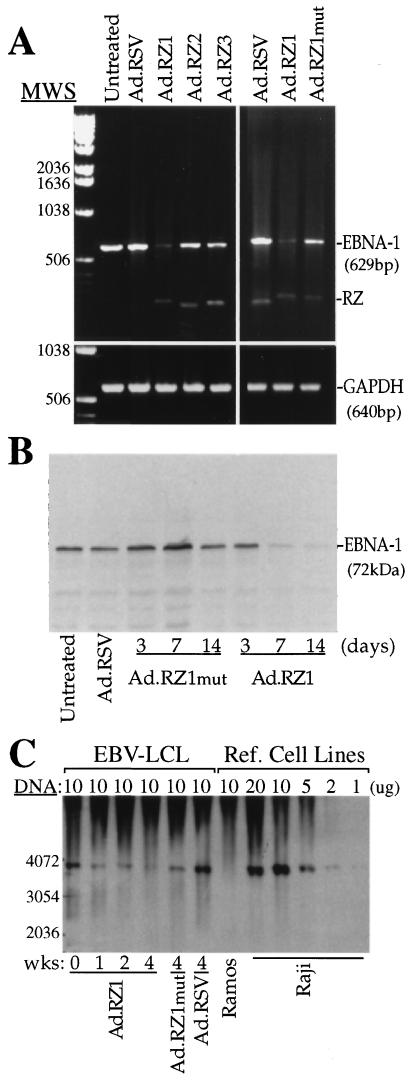
Further studies were performed to determine whether adenovirus-delivered ribozymes could modulate EBNA-1 expression, as well as persistence of the viral genome in B cells previously transformed by EBV. Infection of LCL with Ad.RZ1 significantly reduced EBNA-1 RNA expression while infection with Ad.RZ2 or Ad.RZ3 caused a much smaller reduction in EBNA-1 expression (Fig. 4A). To confirm that the decrease in EBNA-1 RNA expression caused by Ad.RZ1 was due to cleavage of the target RNA, we examined EBNA-1 expression in LCL infected with Ad.RZ1 mut. RZ1 mut can bind EBNA-1 RNA and could potentially exhibit antisense effect but it cannot cleave RNA molecules (Fig. 1). Infection with Ad.RZ1 mut caused only a small decrease in EBNA-1 expression (Fig. 4A) These findings suggest that RZ1 inhibits EBNA expression principally by cleaving EBV RNA.
Figure 4.
Effect of EBNA-1 ribozymes on EBNA-1 expression and EBV genome persistence in LCL cells. (A) EBNA-1 RNA and a control cellular RNA, glyceraldehyde-3-phosphate dehydrogenase, was analyzed by reverse transcription–PCR in LCL 48 hr postinfection with different adenoviruses. (B) Expression of the 72-kDa EBNA-1 protein in LCL infected for varying times with Ad.RZ1 was analyzed by immunoblotting. (C) The presence of the EBV genome in LCL or in EBV-positive (Raji) or EBV-negative (Ramos) was measured by Southern blotting at varying times after adenovirus infection (LCL only) using a 32P-labeled BamHI W fragment.
Further studies examined whether infection of LCL by Ad.RZ1 resulted in suppression of EBNA-1 protein expression. Infection of LCL with Ad.RZ1 resulted in a progressive decrease in EBNA-1 protein expression, whereas infection with Ad.RZ1 mut did not significantly alter EBNA-1 protein expression (Fig. 4B).
To extend these findings, we next examined whether infection of LCL with Ad.RZ1 also reduced persistence of the EBV genome. Infection with Ad.RZ1 resulted in a progressive loss of the EBV genome as indicated by Southern blot analysis (Fig. 4C). This result is consistent with the ability of RZ1 to inhibit EBNA-1 protein expression since maintenance of the EBV genome in latently infected B cells is dependent on expression of EBNA-1.
Adenovirus Delivery of RZ1 Inhibits EBV-Induced Cell Proliferation.
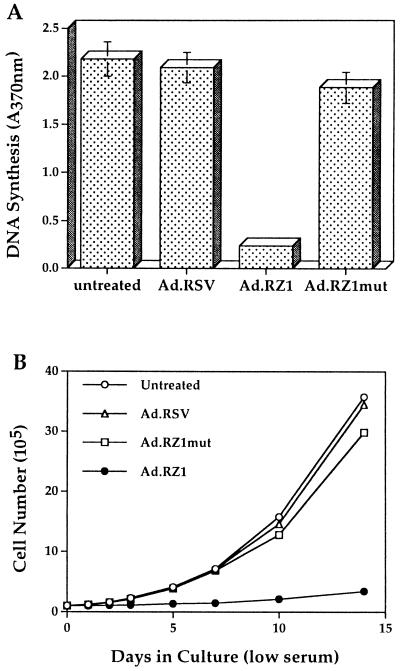
The presence of the EBV genome is required for the initial steps leading to B lymphocyte growth transformation. We therefore investigated whether RZ1-mediated suppression of EBNA-1 expression resulting in the loss of EBV genomes also inhibited LCL proliferation in low serum. Infection with Ad.RZ1 caused a significant reduction in LCL DNA synthesis and also a profound decrease in cell proliferation (Fig. 5). In contrast, infection of LCL with Ad.RZ1 mut caused only a small reduction in DNA synthesis and cell proliferation. Inhibition of cell proliferation by Ad.RZ1 was not due to increased cell death as monitored by trypan blue dye exclusion. These results indicate that EBNA-1 expression and maintenance of the EBV genome are required for cell growth in low serum, and further, that delivery of a specific ribozyme targeted to EBNA-1 can disrupt this process.
Figure 5.
Effect of adenovirus-delivered EBNA-1 ribozymes on LCL cell proliferation. (A) DNA synthesis in either untreated or adenovirus infected LCL cells was detected with an ELISA. (B) The total number of control or adenovirus-infected LCL cells was determined at varying times after culturing in low serum.
DISCUSSION
EBV immortalization of human B lymphocytes involves the expression of multiple gene products (3, 9). Although the pattern of viral gene expression varies from in vitro transformed B cells to EBV-positive human tumors, EBNA-1 is the only viral latent gene product that is consistently expressed (26). This pattern of gene expression reflects the crucial role of EBNA-1 in EBV genome persistence and viral DNA replication. EBV-transformed cells that express only EBNA-1 can avoid recognition by the host immune system since this viral protein is not properly processed (27). The EBNA-1 protein may also have an oncogenic role in the absence of other EBV gene products (12, 13). For these reasons, we sought to determine whether antiviral approaches targeting the EBNA-1 protein could inhibit EBV-induced cell proliferation. Previous studies showed that antisense oligonucleotides directed to EBNA-1 partially inhibited EBNA-1 expression and also suppressed B cell proliferation (28, 29). However, sufficient quantities of antisense oligonucleotides cannot be efficiently delivered to B cells and their relatively short half-life inside cells has hindered utilization of this antiviral approach. To overcome these problems, we investigated whether recombinant adenovirus could be used to efficiently deliver EBNA-1 specific ribozymes. True ribozymes have the potential for enhanced antiviral activity by virtue of their ability to hydrolyze multiple copies of specific RNA molecules. Our studies demonstrated that freshly isolated B cells were relatively resistant to adenovirus infection, whereas EBV-transformed cells expressed αv integrins and were also susceptible to adenovirus gene delivery (Fig. 2). Recent studies have shown that αv integrins are also up-regulated on chronic lymphocytic leukemia B cells by CD40 cross-linking and that this leads to enhanced susceptibility to adenovirus gene delivery (30). Each of the synthetic EBNA-1 ribozymes investigated in these studies was capable of cleaving RNA in vitro, however only RZ1 down-regulated RNA and protein expression in LCL cells (Fig. 4). The failure of RZ2 and RZ3 to modulate EBNA-1 expression in cells could be due to the fact that the predicted cleavage/binding sites recognized by these ribozymes are not accessible in vivo (20). RZ1 also greatly decreased the amount of EBV genomes in LCLs and suppressed cell growth in low serum (Figs. 4 and 5). Previous studies have shown that the proliferation of LCLs at early passages is critically dependent upon the presence of high serum concentrations, cell feeder layers, or cell density (31). The spontaneous loss of the EBV genome in a Burkitt lymphoma cell line, Akata, prevented proliferation of these cells in low serum and also inhibited their tumorigenicity in nude mice (32). These features reflect the involvement of specific cytokines such as interleukin 5, interleukin 6, interleukin 10, and tumor necrosis factor-β that comprise autocrine pathways that are required for EBV-induced B cell proliferation (33).
Although the present studies demonstrate that an EBNA-1 specific ribozyme (RZ1) can inhibit EBV-induced cell proliferation in vitro, it remains to be determined whether similar results can be achieved in vivo. Approximately 10-fold higher amounts of recombinant adenovirus vector are required to efficiently transduce EBV-transformed B cells relative to that needed for epithelial cells, a common cell type infected by adenovirus. Thus further studies are needed to improve this situation for potential in vivo applications. It is worth noting that the RZ1 target sequence is conserved in the EBNA-1 protein of different EBV strains (34) thus increasing the likelihood that this ribozyme can be effective against multiple EBV types in vivo. Moreover, adenovirus vectors may be well suited for delivering antiviral agents for EBV-induced diseases in immunocompromised patients since the elimination of adenovirus-transduced cells by the host immune system should be diminished (35, 36). In addition to treating lymphoproliferative disorders, adenovirus-delivered EBNA-1 ribozymes could potentially be used for EBV-associated diseases involving infection of epithelial cells, nasopharyngeal carcinoma, Sjogren syndrome, and oral hairy leukoplakia in AIDS patients (3). Independent of their use in antiviral therapy, EBV-specific ribozymes are also valuable tools with which to investigate the precise events involved in EBV-induced B cell activation, transformation, and viral replication.
Acknowledgments
We thank Dr. Lindsay Whitton for advice on constructing the EBNA-1 ribozymes. This work was supported by National Institutes of Health Grants CA36204, HL54352, and 2MO1 RR00833, and an American Cancer Society Fellowship to S.H.
ABBREVIATIONS
- EBV
Epstein–Barr virus
- EBNA-1 and -2
EBV nuclear antigen 1 and 2
- LCL
lymphoblastoid cell lines
- LMP-1 and -2
latent membrane protein 1 and 2
- GFP
green fluorescent protein
- RSV
Rous sarcoma virus
References
- 1.Klein G, Klein E. Nature (London) 1985;315:190–195. doi: 10.1038/315190a0. [DOI] [PubMed] [Google Scholar]
- 2.Henle G, Henle W, Diehl V. Proc Natl Acad Sci USA. 1968;59:94–101. doi: 10.1073/pnas.59.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rickinson A B, Kieff E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 2397–2446. [Google Scholar]
- 4.Penn I. Transplant Proc. 1979;11:1047–1051. [PubMed] [Google Scholar]
- 5.Nakanishi M, Kikuta H, Tomizawa K, Kojima K, Ishizaka A, Okano M, Sakiyama Y, Matsumoto S. Cancer. 1997;72:1376–1381. doi: 10.1002/1097-0142(19930815)72:4<1376::aid-cncr2820720437>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 6.Moss D J, Rickinson A B, Pope J H. Int J Cancer. 1979;23:618–625. doi: 10.1002/ijc.2910230506. [DOI] [PubMed] [Google Scholar]
- 7.Misko I S, Soszynski T D, Kane R G, Pope J H. Clin Immunol Immunopathol. 1984;32:285–297. doi: 10.1016/0090-1229(84)90273-3. [DOI] [PubMed] [Google Scholar]
- 8.Moss D J, Rickinson A B, Pope J H. Int J Cancer. 1978;22:662–668. doi: 10.1002/ijc.2910220604. [DOI] [PubMed] [Google Scholar]
- 9.Kieff E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 2343–2396. [Google Scholar]
- 10.Cohen J I, Wang F, Mannick J, Kieff E. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaye K M, Izumi K M, Kieff E. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivas S K, Sixbey J W. J Virol. 1995;69:8155–8158. doi: 10.1128/jvi.69.12.8155-8158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson J B, Bell J L, Levine A J. EMBO J. 1996;15:3117–3126. [PMC free article] [PubMed] [Google Scholar]
- 14.Yates J, Warren N, Reisman D, Sugden B. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlins D R, Milman G S, Hayward S D. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 16.Reisman D, Sugden B. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugden B, Warren N. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W. Current Protocols in Immunology. New York: Wiley; 1992. pp. 7.5.1–7.5.3. [Google Scholar]
- 19.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W. Current Protocols in Immunology. New York: Wiley; 1992. pp. 7.22.1–7.22.2. [Google Scholar]
- 20.Bratty J, Chartrand P, Ferbeyre G, Cedergren R. Biochim Biophys Acta. 1993;1216:345–359. doi: 10.1016/0167-4781(93)90001-t. [DOI] [PubMed] [Google Scholar]
- 21.Becker T C, Noel R J, Coats W S, Gomez-Foix A M, Alam T, Gerard R D, Newgard C B. Methods Cell Biol. 1994;43:161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 22.Lieber A, He C-Y, Polyak S J, Gretch D R, Barr D, Kay M A. J Virol. 1996;70:8782–8791. doi: 10.1128/jvi.70.12.8782-8791.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S, Endo R I, Nemerow G R. J Virol. 1995;69:2257–2263. doi: 10.1128/jvi.69.4.2257-2263.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 25.Stupack D G, Shen C, Wilkins J A. Exp Cell Res. 1992;203:443–448. doi: 10.1016/0014-4827(92)90019-5. [DOI] [PubMed] [Google Scholar]
- 26.Rowe M, Rowe D T, Gregory C D, Young L S, Farrell P J, Rupani H, Rickinson A B. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Nature (London) 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 28.Roth G, Curiel T, Lacy J. Blood. 1994;84:582–587. [PubMed] [Google Scholar]
- 29.Pagano J S, Jimenez G, Sung N S, Raab-Traub N. Ann NY Acad Sci. 1992;660:107–116. doi: 10.1111/j.1749-6632.1992.tb21063.x. [DOI] [PubMed] [Google Scholar]
- 30.Cantwell M J, Sharma S, Friedmann T, Kipps T J. Blood. 1996;88:4676–4683. [PubMed] [Google Scholar]
- 31.Klein G. In: Advance in Viral Oncology. Klein G, editor. New York: Raven; 1987. pp. 207–211. [Google Scholar]
- 32.Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, Takada K. J Virol. 1994;68:6069–6073. doi: 10.1128/jvi.68.9.6069-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon J, Ley S C, Melamed M D, Aman P, Hughes-Jones N C. J Exp Med. 1984;159:1554–1559. doi: 10.1084/jem.159.5.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhatia K, Raj A, Gutiérrez M I, Judde J G, Spangler G, Venkatesh H, Magrath I T. Oncogene. 1996;13:177–181. [PubMed] [Google Scholar]
- 35.Yang Y, Nunes F, Berencsi K, Furth E E, Gönczöl E, Wilson J M. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelhardt J F, Ye X, Doranz B, Wilson J M. Proc Natl Acad Sci USA. 1994;91:6196–6200. doi: 10.1073/pnas.91.13.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]