Abstract
Tissue transglutaminase (tTG) is a multifunctional enzyme with transglutaminase crosslinking (TGase), GTP binding, and hydrolysis activities that play a role in many different disorders. We identified, characterized, and investigated the function and stability of two alternatively spliced forms of tTG using biochemical, cellular, and molecular biological approaches. Using a human aortic vascular smooth muscle cells (VSMC) cDNA library, we identified two cDNAs encoding C-terminal truncated forms, tTGV1 and tTGV2. tTGV1,2 mRNAs were synthesized by a rare splicing event using alternate splice sites within exons 12 and 13 of the tTG gene, respectively. Quantitative PCR and immunoblotting demonstrated that there was unique expression and localization of tTGV1,2 compared with tTG in human umbilical vein endothelial cells (HUVECs), VSMC, and leukocytes. The loss of C-terminal 52 amino acid residues (AAs) in tTGV1,2 altered GTP binding, enhanced GTP hydrolysis, rendered the variants insensitive to GTP inhibition, and resulted in <10% residual Ca+2-dependent TGase activity. Transfection in HEK293 demonstrated a 28- and 5-fold reduction in the expression of tTGV1 and tTGV2, respectively, demonstrating that the C-terminal GTP-binding domain is important in stabilizing and promoting the half-life of tTG. The altered affinity for GTP allowed tTGV1,2 to exhibit enhanced TGase activity when there is a transient increase in Ca+2 levels. The abundance of tTGV1,2 and its distinct intracellular expression patterns in human vascular cells and leukocytes indicate these isoforms likely have unique physiological functions.—Lai, T. S., Liu, Y., Li, W., Greenberg, C. S. Identification of two GTP-independent alternatively spliced forms of tissue transglutaminase in human leukocytes, vascular smooth muscle and endothelial cells.
Keywords: neurodegenerative disease, aging, cell differentiation, extracellular matrix, vascular disease
Tissue transglutaminase (tTG) is a unique member of the transglutaminase gene family (TG, E.C. 2.3.2.13, protein-glutamine γ-glutamyltransferase) because it exhibits multiple enzymatic properties. Established first as a Ca+2-dependent transglutaminase enzyme (TGase) catalyzing an isopeptide bond between a specific γ-glutamyl (Q) containing peptide substrate and an ε-amine group from a peptide-bound lysine (K) residue or a free primary amine (1), it was later found to have G protein signaling and kinase activities (2-5). Moreover, tTG has a wide variety of functions independent of its enzymatic activities (6). tTG is known to function as a cell-surface adhesion molecule, to bind nitric oxide (NO), to serve as a coreceptor for integrins and the G-protein coupled receptors (GPCRs), and to be involved in signaling functions (6-11). tTG also has roles in promoting cell death and survival and inflammation and has been identified as the therapeutic target in neurodegenerative, fibrotic, autoimmune, and cardiovascular disorders (6, 12-14). However, a comprehensive understanding of how a single gene product can perform such a diverse array of functions and activities at distinct cellular localizations is lacking. Indeed, the many attributes of tTG suggest that post-translational modifications and/or alternative splicing (15, 16) may act to generate a subfamily of tTG molecules with unique characteristics.
We routinely observed lower molecular mass (m) tTG antigens when proteins were extracted from various human coronary homogenate and wound healing tissues and analyzed by SDS-PAGE and immunoblotting (17, 18). Because tTG is highly expressed in vascular smooth muscle cells (VSMC), we screened a cDNA library derived from human aortic smooth muscle cells to identify whether these tTG antigens were formed by proteolysis or by alternate splicing. We report two novel alternatively spliced forms of tTG and the splicing mechanism that produced these isoforms. We characterized the biochemical properties of these isoforms, and the expression of these isoforms in different cell types. The in situ stability and effects of these isoforms on cell viability and apoptosis were investigated. Finally, we found that these proteins were expressed predominantly in the cytoplasm of human umbilical vein endothelial cells (HUVECs) and in the nuclei of VSMCs and were selectively expressed in human leukocytes.
MATERIALS AND METHODS
Materials
Monoclonal antibodies to human tTG (7402 and TG100) were purchased from Labvision (Fremont, CA, USA). A mouse monoclonal antibody to tTG, cub7401, was obtained as a gift from Dr. Birckbichler (19). Monoclonal antibody 4C1 specific to tTG (20) was purchased from the Hybridoma Bank (University of Iowa). 5-Biotin-amido-pentylamine (BP) was obtained from Pierce (Rockford, IL, USA). Purified plasma fibronectin (FN) was obtained from Calbiochem (San Diego, CA, USA). A monoclonal antibody to poly (ADP-ribose) polymerase (PARP) was purchased from PharMingen (San Diego, CA, USA). All other reagents used in this investigation were purchased from Sigma (St. Louis, MO, USA) unless otherwise noted.
cDNA library screening and DNA sequencing
Up to 1 × 106 plaque forming units (pfu) of lambda phages from the human aortic VSMC lambda ZAP cDNA library were screened using a [32P]-labeled full-length tTG cDNA probe (18) according to procedures recommended by the manufacturer (Stratagene, La Jolla, CA, USA). Positive clones were purified by repeated plating and hybridization. The pBluescript- (pBSK-) carrying cDNA insert was then excised from those that were identified as being positive and were confirmed by PCR amplification using built-in T3 and M13 primers in a pBluescrip vector (Stratagene). The size of the cDNA insert was validated by digesting the cDNA clones with EcoRI and XhoI. DNA sequencing analysis of the cDNA inserts was performed at least twice from both strands of cDNA using overlapped oligonucleotide primers derived from human tTG mRNA (NCBI Accession # NM_004613). The clones containing cDNAs encoding tTGV1 and tTGV2 were designated as pBSK-TTGV1 and pBSK-TTGV2, respectively.
E. coli and mammalian expression
A previously constructed plasmid, pGEX-MCS (18), to express the recombinant protein in E. coli as a glutathione-S-transferase (GST) fusion protein was used as a template for the construction of pG-tTGV1 and pG-tTGV2. The NcoI-KpnI or NcoI-HindIII full-length cDNA fragment of tTGV1 or tTGV2 releasing from pBSK-TTGV1 and pBSK-tTGV2, respectively, were subcloned into the pGEX-MCS vector previously digested with NcoI-KpnIor NcoI-HindIII, to form pG-TTGV1 and pG-tTGV2, respectively To construct the mammalian expression vector, the NcoI-HindIII fragment of full-length tTG cDNA releasing from pG-TG was ligated into the pBK-CMV (Stratagene) vector previously digested with NheI and HindIII. The resulting plasmid DNA was designated as pBKCMV-tTG. To construct pBKCMV-tTGV1 and pBKCMV-tTGV2, AvaI-KpnI fragments carrying tTGV1 and tTGV2 cDNAs were isolated from pBSK-tTGV1 and pBSK-tTGV2 and used to replace the corresponding restriction fragment in pBKCMV-tTG. The final constructs of E. coli and mammalian expression vectors were verified by restriction mapping and DNA sequencing.
Cell Culture
HL-60 cells from ATCC (Rockville, MD, USA) were maintained under subconfluent conditions in RPMI medium with 15% fetal bovine serum (FBS). Human embryonic kidney 293 cells (ATCC) were maintained in Dulbecco’s modified Eagle’s (DMEM) medium/10% FBS and 100 U/ml penicillin-streptomycin at 37°C, 5% CO2. Human umbilical vein endothelial cells (passages 2-5) and aortic vascular smooth muscle cells (Clonetics, Cambrex; Rockland, MD, USA) were cultured in EGM-2 and SmGM medium, respectively, based on the manufacturer’s recommendations (Clonetics).
Drug treatment
In all experiments, HL-60 cells in the exponential phase of growth (<1×106 cells/ml) were used for trans-retinoic acid (RA) treatment (Sigma). Cells were diluted to 5 × 105/ml in fresh RPMI medium with complete serum and treated with either 6 μM RA delivered in a solution of DMSO (vehicle) or in a control vehicle (DMSO) for 16 h.
Expression and Purification of Recombinant tTG, tTGV1, and tTGV2
The recombinant tTG, tTGV1, and tTGV2 were expressed and purified in E. coli as a GST fusion protein as described previously (9). Protein concentrations were quantified using the Bradford method (Bio-Rad, Hercules, CA, USA). The purified proteins in 50 mM Tris acetate buffer, pH 7.5, and 15% glycerol were stored at -80°C until ready for use. The purified proteins were visualized in a Coomassie blue-stained gel and used to normalize the protein concentration.
Transfection of HEK-293 Cells
The HEK-293 cells (ATCC) were transfected with Transfast transfection reagent according to manufacturer’s instructions (Promega, Madison, WI, USA). The transfection efficiency was typically >80-90% as monitored by transfection with vector expressing EGFP. Cells were splitted to 60-80% confluent in a 6-well tissue culture plate 18 h before transfection. pBKCMV-tTG, pBKCM-tTGV1, or pBKCMV-tTGV2 (2 μg) was mixed with Transfast reagent in serum-free DMEM medium at room temperature for 10-20 min before application to the cell monolayer. After incubation at 37°C for 24 h in complete media, the cells were washed 3× with PBS, 0.5 mM EDTA. Cells were scraped from the wells in Tris-buffered saline (50 mM Tris, pH, 7.5, 100 mM NaCl), 0.5% Nonidet P-40, 0.1% sodium cholate, 2 mM EDTA, and protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA) on ice. Cells were sonicated on ice for 2 × 5 s and centrifuged at 10,000 g at 4°C for 20 min to remove the insoluble debris. The soluble fraction was removed and stored at -80°C.
Lactate Dehydrogenase (LDH) Release Assay
A microtiter plate based colorimetric assay (Cytotox 96 nonradioactive cytotoxicity kit; Promega) that measures the release of LDH into cell culture supernatant was used.
Cell Viability assay
A CellTiter-Blue cell viability assay kit (Promega) that measures the ability of live cells to reduce the resazurin to a highly fluorescent compound, resorufin, which can be easily quantified in a fluorescent plate reader (579ex/584em), was used.
Cell Based Caspase 3/7 Activation assay
The activation of caspases 3/7 was measured using a Apo-One homogeneous caspases-3/7 assay kit (Promega). Briefly, cells were lysed and the caspases cleavage activity was determined using Z-DEVD-rhodamine110. When Z-DEVD-R110 is cleaved, an intense fluorescent can be measured in a fluorescent plate reader (499ex/521em).
Preparation of cell lysates from cytoplasmic and nuclear fractions
HUVECs and VSMCs at their exponential growth phase were detached from the plate using trypsin. Cell pellets were obtained by slow speed centrifugation and lysed using NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Rockford, IL, USA) in the presence of a protease inhibitor cocktail (Roche Applied Science). The cytoplasmic and nuclear fractions were prepared according to manufacturers’ instructions. The protein concentrations were determined by the micro BCA method (Pierce). Immunoblot analysis of the nuclear poly (ADP-ribose) polymerase (PARP) protein was used as a marker to validate correct nuclear fractions were isolated. A cell fractionating kit (Qiagen, Valencia CA, USA) was used to separate the lysates into cytosolic, membrane, nuclear, and cytoskeletal fractions.
Preparations of mononuclear leukocyte lysates
Ten milliters of whole blood were donated by health individuals and collected in EDTA-treated tubes. Mononuclear cells (MNs) containing monocytes and lymphocytes were isolated by density gradient centrifugation using Ficoll-Paque Plus solution (GE HealthCare, Piscataway, NJ, USA), with minor modification to remove platelets (21). Briefly, anticoagulant-treated whole blood was centrifuged at 180 g for 10 min and the upper phase containing platelet-rich plasma (PRP) was removed. Cells from the bottom phase were mixed 1:1 with Hanks’ balanced salt solution (without Ca+2 and Mg+2; Invitrogen, Carlsbad, CA, USA) and carefully layered onto an equal volume of Ficoll-Paque Plus solution and spun at 400 g for 40 min at room temperature. Based on manufacturer’s protocol (GE HealthCare), 95% of the isolated cells were peripheral MNs. Total lysates were immediately prepared by resuspending the cell pellets in TBS, 1 mM EDTA, and a protease inhibitor, and then sonicated and stored at -80°C.
TGase Assay
TGase activity was determined by quantifying the incorporation of 5-biotinamidopentylamine (BP) into N,N’-dimethylcasein in a microtiter plate as described previously (22). The concentrations of affinity purified tTG, tTGV1, and tTGV2 were normalized using Coomassie blue-stained SDS-PAGE gel to ensure that equal amounts of each protein was used. For GTP inhibition, GST-tTG (1 μg/ml), GST-tTGV1 (16 μg/ml), or GST-tTGV2 (16 μg/ml) was incubated with different concentrations of GTP in triplicate in the presence of 1 mM CaCl2, and the amount of BP incorporated into the casein was determined after a 40 min incubation at 37°C and color developed using streptavidin-conjugated alkaline phosphatase and PNPP (22).
Fluorescence GTP Binding Assay
The binding assay was performed using a fluorescent GTP analog, BODIPY FL-GTP (Molecular Probes, Invitrogen) essentially as described previously (23, 24). All fluorescence measurements were performed in a cuvette and determined in a Molecular Devices (Sunnyvale, CA, USA) SpectraMax M2e fluorescence reader. The experiments were carried out in 50 mM Tris-HCl buffer (pH 7.5) containing 2 mM DTT and 1 mM EDTA. The detection for BODIPY FL GTP was set at λex 485 nm and λem 520 nm with a λ495nm cut-off.
γ-[P32]-GTP Hydrolysis
The GTP hydrolysis was performed by measuring the release of free phosphate (Pi) from γ-[P32]-GTP (PerkinElmer) as described previously (9).
Trypsin Proteolysis of Recombinant tTG, tTGV1, and tTGV2
Purified recombinant GST-tTG, GST-tTGV1, or GST-tTGV2 (1 μg) was incubated with 0.1 μg of trypsin [l-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK)-treated and high pressure liquid chromatography-purified; Calbiochem Biosciences, San Diego, CA, USA] in the presence of 10 μM free GTP and incubated at 37°C for 1 h. The reaction was stopped by the addition of SDS-PAGE loading buffer. Samples were separated by SDS-PAGE and analyzed by immunoblotting using the monoclonal antibody CUB7402 against human tTG.
FN Binding
A 96-well plate coated with FN binding was used to measure the binding of tTG, tTGV1, and tTGV2, as described previously (25). The bound tTG and isoforms were detected using monoclonal antibody cub 7401, and peroxidase conjugated goat anti-mouse IgG and color was developed using TMB substrate. The color was recorded using OD490nm.
SDS-PAGE and Immunoblotting
Different amounts of cell lysates or purified recombinant proteins were dissolved in 1× SDS-PAGE loading buffer and separated by 8% SDS-PAGE electrophoresis. After gel electrophoresis (PAGE) and transfer to a nitrocellulose membrane, the bands were detected using monoclonal antibody against tTG (Labvision) and visualized using Super Signal West Pico Luminol (Pierce).
RNA isolation
To ensure that processed mRNAs were used, total cytoplasmic RNAs were prepared using an RNeasy kit (Qiagen) according to the manufacturer’s instructions. The DNase-treated total RNAs used in this study showed no signs of degradation, as validated by the presence of intact 18S and 28S rRNA bands using denaturing agarose gel electrophoresis. Peripheral blood total RNAs from human mononuclear cells (MNs) and neutrophils were purchased from AllCell, LLC (Berkeley, CA, USA). Human peripheral blood MNs and neutrophils used to isolate RNAs were >95% in purity based on FACScan analysis report supplied by the manufacturer (AllCell).
Quantification of mRNAs encoding tTG, tTGV1, and tTGV2
Real-time TaqMan PCR assays were performed using either the ABI 7900 system (Applied Biosystems, Foster City, CA, USA) or the Bio-Rad iQ5 system based on the 5′ nuclease assay first described by Holland et al. (26). It uses the 5′-3′ exonuclease activity of TaqMan DNA polymerase to cleave a dual-labeled probe (5′-end FAM as a reporter dye; emission λmax=518 nm and 3′-end TAMRA as a quencher; emission λmax=582 nm) annealed to the target sequence during PCR amplification. The PCR cycle at which fluorescence reached a threshold value 10 times that of the sd of baseline emission [CT, cycle of threshold] was used for the quantitative measurements. Samples of total RNAs were reverse-transcribed using oligo(dT) and Superscript reverse transcriptase (II) followed by RNase H digestion (Invitrogen) according to the manufacturer’s instructions. The synthesized cDNAs were amplified using a TaqMan universal PCR Master Mix (Applied Biosystems). Amplicon size and reaction specificity were confirmed by agarose gel electrophoresis (see supporting figures). The following TaqMan PCR assay primers and probes were designed by Applied Biosystem’s custom assay-by-design service. The oligonucleotide sequences were verified for specificity with the NCBI’s Blast engine using the “nearly exact short match” program. Each TaqMan assay was designed to cross the exon-exon junction to differentiate premessanger (premRNA) and genomic DNA contamination and specifically target each tTG, tTGV1, and tTGV2 mRNA (see Fig. 1). The following forward, reverse, and TaqMan probes were used: tTG: forward 5′GGCCGGCCTGACTGA 3′, reverse 5′ AGGTCCATTCTCACCTTAACTTCCT-3′, probe, FAM5′-AAGACGGTGGAGATCCCAGAC-3′TAMRA, base pairs amplified 90 bp; tTGV1: forward 5′-TCCCTGTGGCCCTGGAA-3′, reverse 5′CACATAGTAGGTGCTTCACAATGGT-3′, probe FAM 5′-CTGGGATGTGGAGGTGC-3′ TAMRA, base pairs amplified 99 bp; tTGV2: forward 5′-TCCCTGTGGCCCTGGAA-3′, reverse 5′-TTCACATTACCCAGCCTTGCTAA-3′, probe FAM 5′-CTGCACCTCCACACCAGT-3′ TAMRA, base pairs amplified 75 bp.
Figure 1.
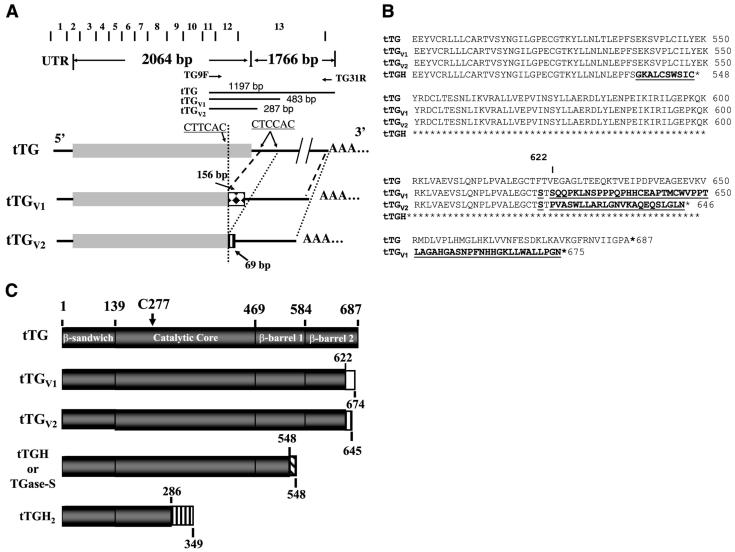
Alignment of tTG, tTGV1 and tTGV2 cDNAs and encoded protein sequences. A) Alignment of cDNAs encoding tTG, tTGV1, and tTGV2 along with exon-intron junctions. The 5′-UTR, coding region, and 3′ UTR region are illustrated along with the exon-exon junction of tTG. Gray closed box represents coding region of wild-type tTG. When PCR was used to amplify cDNA prepared from HUVEC total RNAs using TG9F and TG31R primer pairs, DNA fragments of 1197, 483, and 287 bp were amplified, representing mRNA encoding tTG, tTGV1, and tTGV2, respectively. B) Alignment of translated protein sequences of tTG, tTGV1, and tTGV2. Amino acid 622 is marked to show location of different amino acid sequences in tTGV1 and tTGV2. Different AAs in tTGV1 and tTGV2 are bolded and underlined. 1C. Schematic diagram of tTG and different isoforms. Gray box represents tTG protein sequences, and open box and other shaded boxes represent alternate amino acid sequences due to change in reading frames.
Amplification was performed using a 2 min 50°C and 95°C for 10 min activation, followed by 45 two-step cycles (15 s 95°C, 1 min 60°C) on a 2 μl cDNA (corresponding to 30 ng total RNA) with 1× TaqMan Universal PCR Master Mix, 900 nM each of forward and reverse primer, and 250 nM Taqman probe at a total volume of 25 μl. Serial diluted plasmid DNA (pBKCMV-tTG, tTGV1, or tTGV2) was amplified in the same plate and used to establish a standard calibration curve to calculate the absolute copy number of the tTG, tTGV1, and tTGV2 mRNAs. To control the variation in the amount of cDNA available for PCR in the different samples, gene expression of the target sequence was normalized in relation to the expression of an endogenous control, human β-actin (Applied Biosystems). Copy number was calculated using the following formula: A × 6.023 × 1023/(B) × 330 × 2; A: DNA amount in gm; B: DNA length in basepairs.
Molecular Modeling
The three-dimensional coordinates of the human tTG are available from the Protein Data Bank (Research Collaboratory for Structural Bioinformatics, Rutgers University) with access code 1kv3 (27). The detailed interaction map of AAs within tTG was obtained from the Protein Families Database of Alignments and HMMs (pfam) (www.sanger.ac.uk/software/pfam) and Simple Modular Architecture Research Tool (SMART at http://smart.embl-heidelberg.de/smart).
RESULTS
Cloning and Identification of two alternatively spliced forms of tTG
When total cell lysates derived from HUVECs and VSMCs were analyzed on an immunoblot using monoclonal antibodies cub7401, cub7402 and TG100 (Labvision, CA), one full-length and two lower molecular mass of tTG-related antigens were routinely observed (see Fig. 1 and Fig. 2). The specificity of these antibodies was validated to react with full-length tTG, tTGV1 and tTGV2 (see Supplemental Materials). To determine whether the truncated forms of tTG were formed by proteolysis or by alternative splicing, we screened a human aortic VSMC lambda ZAP cDNA library (Stratagene) using the entire coding region of human tTG cDNA as a probe. All positive clones were completely sequenced from both DNA strands using overlapping DNA primers derived from published human tTG cDNA (Gene Bank # NM_004613); 57% of the positive cDNA clones encoding full-length tTG (Gene Bank # NM_004613), and two pools of positive cDNA clones (with 14.3% each), encoding for tTGV1 and tTGV2 with internal deletion at 3′-end were identified (Fig. 1). Two of the known tTG isoforms, tTGH (or designated as TGase-S) and tTGH2 (Fig. 1C) (28), were not found in the library screening.
Figure 2.
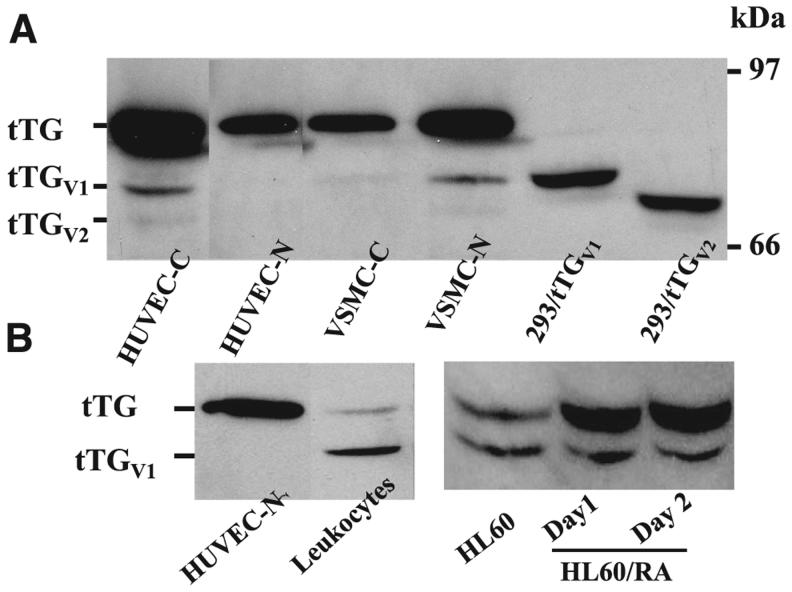
Immunoblot analysis of tTG and its isoforms. A) Immunoblot of HUVECs, VSMCs, and HEK 293 cells transfected with pBKCMV-tTGV1 and pBKCMV-tTGV2. Cytoplasmic (C) and nuclear (N) fractions of cell lysates were prepared from HUVECs and VSMCs (designated HUVEC-C, HUVEC-N, VSMC-C, and VSMC-N, respectively). In addition, total cell lysates were prepared from HEK293 cells transfected with pBKCMV-tTGV1 and pBKCMV-tTGV2. Amount loaded in each lane: HUVEC-C: 1 μg; HUVEC-N: 1 μg; VSMC-C: 1 μg; VSMC-N: 1 μg; 293/pBKCMV-tTGV1: 25 μg; 293/pBKCMV-tTGV2: 25 μg. B) Immunoblot analysis of cell lysates prepared from HUVECs and HL-60 cells. HUVEC-C (1 μg) and total cell lysates prepared from human MNs and HL-60 and HL-60/RA (200 μg each) were separated by SDS-PAGE gel electrophoresis and transferred to a nitrocellulose membrane, and the blot was probed with monoclonal antibody against tTG (cub 7402, Labvision).
DNA sequence analysis revealed that the cDNAs encoding tTGV1 and tTGV2 were produced by alternate splicing events using alternate 5′ and 3′ splice sites located within exons 12 and 13 of tTG (Fig. 1A), respectively. tTGV1 and tTGV2 mRNAs were generated by the deletion of DNA sequences between 1963CT/TCAC and 2682CT/CCA, and 1963CT/TCAC and 2878CT/CCACC (nucleotide number refers to NM_004613;/denotes spliced sites), respectively (Figs. 1A and Supplemental Fig. 1). The existence of mRNAs encoding tTGV1 and tTGV2 were validated by PCR of reverse-transcribed cDNAs synthesized from total cytoplasmic mRNAs. PCR primer pairs flanking the spliced sited were able to amplify 1197 bp (for tTG), 483 bp (for tTGV1), and 287 bp (for tTGV2) cDNA fragments from HUVECs and leukocytes (data not shown). When the open reading frames were analyzed, cDNAs encoding tTGV1 and tTGV2 were composed of 674 and 645 AAs, respectively, that shared 622 identical AAs on the N terminus with wild type tTG, with an alternate 52 and 23 AAs at the C terminus, and translated into proteins with a predicted m of 75 and 70 kDa, respectively (Fig. 1B).
Distinct localization of tTG and isoforms in human MNs and vascular cells
To validate the expression of these isoforms, several antibodies including cub7401, cub7402, TG100, and 4C1 were epitope-mapped using purified recombinant proteins; we found that cub7401, cub 7402, and TG100 antibodies reacted with tTGV1 and tTGV2 (see Supplemental Materials). To validate their expression, cell lysates prepared from HEK293 cells transiently transfected with pBKCMV-tTGV1 and pBKCMV-tTGV2 and tTGV1 and tTGV2 could be easily detected on an immunoblot (Fig. 2A). We also analyzed lysates prepared from cytosolic and nuclear fractions of HUVECs and VSMCs. To ensure that the correct fractions were isolated, immunoblots using monoclonal anti-PARP demonstrated that PARP was only detected in nuclear fractions (data not shown). With the same amount of total lysates, untransfected HEK 293 cells expressed undetectable tTG-related antigen. When equal amount of cell lysates isolated from cytosolic and nuclear fractions of HUVECs and VSMCs were analyzed (Fig. 2A), tTG was the major form in the cytosolic fractions of HUVEC, and about the same amount of tTG was detected in both the cytosolic and nuclear fractions of VSMC cells (Fig. 2A). Interestingly, the tTGV1 and tTGV2 were detected predominately in the cytosolic fractions of HUVECs and the nuclear fractions of VSMCs, respectively (Fig. 2A). In cytosolic fractions of HUVECs, tTG was expressed at 10- and 60-fold higher levels than tTGV1 and tTGV2, respectively (Fig. 2A). Similarly, tTG was expressed at 7- and 45-fold higher levels than tTGV1 and tTGV2 in nuclear fractions of VSMCs, respectively (Fig. 2A). When cell lysates from human MNs were analyzed, tTGV1 was expressed at ∼10 fold higher levels than tTG, but tTGV2 was not detected (Fig. 2B). RA treatment of HL-60 cells served as the positive control as it is known to induce tTG (Fig. 2B). After overnight RA treatment, tTG was induced at least 10-fold, but tTGV1 was not changed significantly (Fig. 2B).
Differential regulation of mRNAs encoding tTG and isoforms
In an effort to validate the expression of mRNAs encoding tTG, tTGV1, and tTGV2, specific TaqMan PCR assays were designed to quantify the expression of different isoforms according to the procedures described in Materials and Methods. TaqMan assays were used because of their specificity, requiring the binding of PCR primer pairs plus the fluorescent probe primer to generate a fluorescent signal.
Table 1 summarizes the results from real-time TaqMan assays. In HUVEC cells, tTG mRNA was expressed at 32- and 194-fold higher levels than tTGV1 and tTGV2 mRNAs, respectively (Table 1). The expression of tTG and tTGV1/tTGV2 mRNAs of MNs and PMNs isolated from healthy individuals was also investigated (Table 1). In human MNs, the tTG mRNA was expressed at 4.4- and 5.3-fold higher levels than tTGV1 and tTGV2 mRNAs, respectively, while tTG mRNA in PMNs was expressed at 0.5- and 2.3-fold higher levels than tTGV1 and tTGV2 mRNAs, respectively (Table 1). In HL-60 cells, a promyelocytic leukemia cell line, the expression of mRNA encoding tTG was 28- and 2 fold higher than that of tTGV1 and tTGV2 mRNAs (Table 1). RA induced the tTG, tTGV1, and tTGV2 mRNAs 3000-, 2000-, and 21-fold, respectively, in HL-60 cells.
TABLE 1.
Expression of mRNAs encoding tTG, tTGV1, and tTGV2
| tTG | tTGV1 | tTGV2 | tTG/tTGV1 | tTG/tTGV2 | |
|---|---|---|---|---|---|
| HUVEC | 1036,954 ± 46,664 | 32,244 ± 1404 | 5334 ± 322 | 32 | 194 |
| MN | 2486 ± 554 | 569 ± 190 | 465 ± 138 | 4.4 | 5.3 |
| PMN | 52 ± 22 | 114 ± 91 | 23 ± 9 | 0.5 | 2.3 |
| HL-60 | 228 ± 48 | 8 ± 1 | 109 ± 8 | 28.5 | 2 |
| HL-60/RA | 75,2875 ± 26,473 | 15,948 ± 797 | 2305 ± 140 | 47 | 326 |
Data are presented as relative copy number in 30 ng cDNA as measured by specific Taqman probe using a Bio-Rad’s IQ5 real-time PCR instrument. Every measurement was determined by at least 3 duplicate experiments.
When the mRNA expression was expressed as the ratio of tTG/tTGV1 and tTG/tTGV2, there was dramatic difference between HUVECs and human MNs and PMNs (Table 1). In human MNs, PMNs, and HL-60 cells, tTG was expressed at a much lower level than in HUVECs, with tTG/tTGV1 ratios of 4.4, 0.5, and 28.5 for human MNs and PMNs and HL-60 cells, respectively. RA-treatment of HL-60 cells increased the tTG/tTGV1 and tTG/tTGV2 ratio to a level closer to that of HUVECs (Table 1).
tTGV1 and tTGV2 displayed TGase activity and were not inhibited by GTP-γ-S
To determine whether the loss of C-terminal residues had an effect on overall enzymatic activity, the TGase activity of the tTG isoforms was determined using purified proteins and a BP assay as described previously (22). The results showed that tTGV1 and tTGV2 required the same Ca+2 concentrations for optimum activation as tTG but had only 8 and 7% of the TGase activity when the same amount of proteins were used in the assay (Table 2).
TABLE 2.
Specific transglutaminase activity of different isoforms of tTG
| tTG | tTGV1 | tTGV2 | |
|---|---|---|---|
| TGase activity (mOD/min/μg) | 2471 ± 422 | 191 ± 3.9 | 176 ± 8.6 |
TGase assay: Assay was performed measuring BP-incorporated into N,N’-dimethylcasein coated microtiter plate. Activity was average of 2 triplicate experiments.
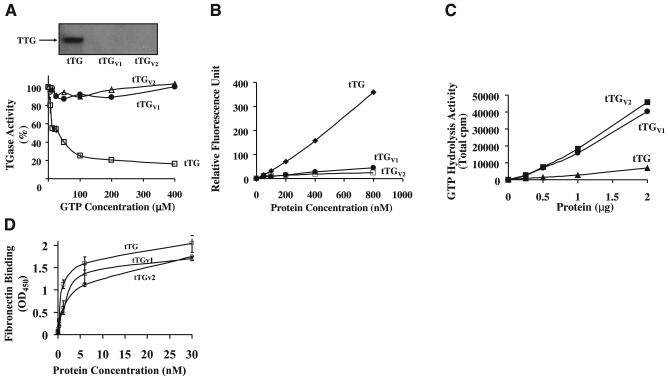
We next examined the effects of GTP on TGase activity of tTGV1 and tTGV2 (Fig. 3). GTP binds to tTG, inducing a conformational change and dramatically inhibiting TGase activity (2, 9). We have reported previously that GTP inhibits tTG with an IC50 ∼6 μM (9). In contrast, tTGV1 and tTGV2 did not show any inhibition by GTP at concentrations up to 400 μM. The conformational change in tTG, tTGV1, and tTGV2 induced by GTP-γ-S was also investigated (Fig. 3, inset). For tTG, GTP-γ-S was able to bind and protect the protein from proteolysis by trypsin. GTP-γ-S showed no protective effect on tTGV1 and tTGV2 under identical conditions, as these isoforms were completely degraded after incubation with trypsin (Fig. 3, inset).
Figure 3.
Effects of GTP on TGase activity and GTP binding of tTG, tTGV1, and tTGV2. A) Effects of GTP-γ-S on TGase activity of tTG, tTGV1, and tTGV2. TGase activity of purified tTG (1 μg/ml), tTGV1 (16 μg/ml), or tTGV2 (16 μg/ml) were determined using BP incorporation assay in reaction buffer containing 0.1 M HEPES buffer, pH 7.5, 1 mM Ca+2, and different concentrations of GTP-γ-S (0-400 μM) at 37°C for 40 min, as described in Materials and Methods. Inset is immunoblot of the trypsin protection assay for tTG and its isoforms. In trypsin protection assay, GST-tTG, -tTGV1, or -tTGV2 (2 μg each) was incubated with 100 μM GTP-γ-S and 50 ng trypsin in 0.1 M Tris-Cl, pH 7.5, for 30 min at 37°C. Samples were separated by SDS-PAGE gel electrophoresis and transferred onto a nitrocellulose membrane. tTG-related antigen(s) was detected using monoclonal antitTG (Cub7402). B) Fluorescent GTP Binding. Fluorescence binding assay was perform in buffer B (50 mM Tris-Cl, pH 7.5, 2 mM DTT and 1 mM EDTA) containing 0-800 nM of purified enzyme (tTG, tTGV1, or tTGV2) and 500 nM BODIPY FL-GTP. Buffer B contained only BODIPY FL-GTP was used as a reference; results represent difference in fluorescence on binding to enzymes. C) GTP hydrolysis. GTP hydrolysis was performed using 0 to 2 μg of purified tTG, tTGV1 and tTGV2 in a reaction buffer containing 50 mM Tris-Cl, pH 7.5, 4 mM Mg+2, 2 mM DTT, 0.1 mM EDTA, [32P]-γ-GTP, and 10 μM unlabeled GTP at 37°C for 30 min. Release of free phosphate was determined using scintillation counting as described previously (Lai, T. S., et al., 1998). D) Fibronectin binding. FN bindings were performed as described under Methods. Data represent 2 independent triplicate determinations.
The altered GTP binding properties were validated using a fluorescent GTP analog, BODIPY-FL-GTP (29). As shown in Fig. 3C, there was a dramatic difference in the enhancing fluorescence intensity on binding to tTG and the isoforms. The increase in fluorescence intensity on binding to tTG was 8-fold stronger than tTGV1 and tTGV2, suggesting that the nucleotide binding pockets in tTGV1/tTGV2 were different from that in tTG (Fig. 3B).
The GTP hydrolysis activity of tTG, tTGV1, and tTGV2 was also investigated (Fig. 3C). The altered nucleotide binding pocket in tTGV1/tTGV2 resulted in a 6- to 7-fold increase in GTP hydrolysis activity.
The ability of tTG, tTGV1, and tTGV2 to bind FN was investigated as described under Materials and Methods. As shown in Fig. 3D, the loss of C terminus in tTGV1 and tTGV2 did not significantly reduced their ability to bind FN.
C terminus of tTG promotes stability and half-life of tTG
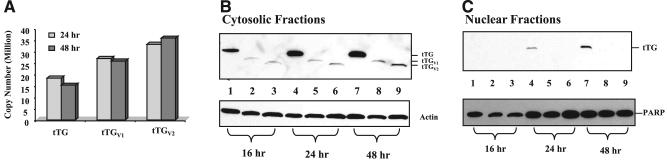
HEK293 cells were transfected with pBKCMV-tTG, pBKCMV-tTGV1 and pBKCMV-tTGV2, and the total RNAs and cellular proteins were isolated 16, 24, and 48 h after transfection to determine the mRNA expression levels and proteins (Fig. 4). Specific TaqMan real-time PCR assays were used to determine the mRNAs encoding tTG, tTGV1, and tTGV2 (Fig. 4A). At the mRNA level, there were 6000-, 7500-, and 9000-fold increases in tTG, tTGV1, and tTGV2, respectively, 24 and 48 h after transfection when compared with untransfected cells (Fig. 4A). At the protein level, the expression of tTG, tTGV1, and tTGV2 in cytosolic fractions reached their maximum levels 48 h after transfection (Fig. 4B). When the intensities of individual protein bands in the X-ray film were quantified, tTG was expressed at 28- and 9-fold higher levels than tTGV1 and tTGV2, respectively (Fig. 4B). tTG was detectable in nuclear fractions 24 and 48 h after transfection, while tTGV1 and tTGV2 were not detected.
Figure 4.
Expression of tTG, tTGV1, and tTGV2 in HEK293 cells. A) Quantitative PCR analysis of mRNAs encoding tTG, tTGV1, and tTGV2. Transfections were performed as described in Materials and Methods, and total RNAs were isolated 24 and 48 h after transfection. Real-time PCR assays were performed using specific Taqman assay. β-actin was used as an internal control to normalize sample loading. B) Immunoblot analysis. Total cell lysates were isolated 16, 24, and 48 h after transfection and fractioned into cytosolic (B) and nuclear fractions (C) as described in Materials and Methods. Lanes 1, 4, 7: cells were transfected with pBKCMV-tTG. Lanes 2, 5, 8: cells were transfected with pBKCMV-tTGV1. Lanes 3, 6, 9: cells were transfected with pBKCMV-tTGV2. In cytosolic fractions, 5 μg of proteins were loaded onto each lane. For the nuclear fractions, 2.5 μg of protein lysates were loaded onto each lane.
Expression of tTG isoforms on cellular viability and caspases 3/7 activation
HEK293 cells were transfected with plasmid vectors, pBKCMV-tTG, -tTGV1 and -tTGV2, and cells were analyzed post-transfection for LDH release, viabili,ty and caspases 3/7 activation according to the procedures described in Materials and Methods (see Supplemental Fig. 5A, B, C). We found that the expression of tTG and isoforms did not have significant effects on LDH release, viability, and caspases activation.
DISCUSSION
We routinely detected lower m form of tTG antigens after SDS-PAGE and immunoblotting in those lysates prepared from tissue culture cell samples and extracts from human atherosclerosis tissues and tissues derived from wounds (17, 18), despite the addition of various protease inhibitors and lowering the temperature during the isolation procedures. The experiments reported here were designed to establish whether these lower m tTG antigens could be formed by alternative splicing in vivo. A combination of molecular, cellular, and biochemical methods demonstrated that there were two unique human C-terminal truncated forms of tTG that localized predominantly to the cytoplasm of HUVECs and the nucleus of VSMCs and were expressed and regulated in a tissue-specific manner. The loss of the native C-terminal sequence resulted in a reduction of Ca+2-dependent TGase activity and in situ stability of the proteins. However, the splice variants had enhanced GTP hydrolysis activity, were active in the presence of GTP, retained FN binding capacity, and were sensitive to proteolysis. Expression of these isoforms in HEK 293 cells did not alter significantly LDH release, viability, and caspases activation suggesting preservation of the antiapoptotic function of tTG by the isoforms (13, 30).
Alternative splicing of premRNA is a fundamental mechanism to increase protein diversity from a single gene (31); 35-59% of human genes have alternative spliced variants, and it is estimated ∼15% of disease-causing mutations in human genes involve mis-regulation of alternative splicing (31). tTGV1 and tTGV2 were identified as the alternate spliced variants of tTG based on the following data: 1) cDNAs encoding tTGV1 and tTGV2 were obtained from a cDNA library constructed from polyadenylated mRNAs; 2) RT-PCR of processed cytoplasmic RNAs using primers flanking the spliced sites were able to amplify three different transcripts with predicted mRNA size encoding tTG, tTGV1, and tTGV2; and 3) tTGV1 and tTGV2 in lysates isolated from HEK293 cells transiently transfected with pBKCMV-tTGV1 and pBKCMV-tTGV2 had the same mobility through an SDS-PAGE gel as those in lysates from HUVECs and VSMCs (Fig. 2). These data excluded the possibility that the tTGV1 and tTGV2 mRNAs were derived from unprocessed premRNAs.
Investigation of the splicing mechanism demonstrated that tTGV1/tTGV2 mRNAs were generated through a rare and specific alternate splicing event that used the combination of alternative 5′ and 3′ spliced sites within exons 12 and 13, respectively (Fig. 1A). The 5′ spliced site CT/TCAC and the 3′ spliced site CT/CCA (/denotes the spliced location) did not match the general consensus sequences for typical splicing (31), i.e., the invariant dinucleotides at each end of the intron, GT at the 5′ end of the intron, and AG at the 3′ end of the intron, and generally resemble MAG/GTRAGT at the 5′ splice site and CAG G at the 3′ splice site (31). The commonly accessed splice site prediction program run by Neural Network (http://www.fruit fly. org/seq_tools/splice.html) did not predict the spliced sites for tTGV1 and tTGV2 mRNAs.
Together with two previously identified isoforms, tTGH and tTGH2 (15, 28; Fig. 1C), there are a total of four human tTG isoforms. The difference is that tTGH and tTGH2 mRNAs are generated by common alternative splicing events through incorporation of introns X and VI (the intron retention mechanism), respectively (31), and encode proteins with 548 and 349 AAs (15, 28). However, these two forms were not detected in our cDNA library screening and may reflect changes derived from leukemogenesis (15). However, tTGH, sometimes referred to as the short isoform (tTG-S or TGase-S) of tTG in the literature (19, 32, 33), was shown to be up-regulated in Alzheimer’s disease (AD) brains, and it is unclear which brain cells produced this protein. A splicing event similar to the one as tTGV1 and tTGV2 mRNAs has also been reported in rat brain astrocytes (referred to as S-TGN; ref 16, 34). Rat S-TGN was induced together with full-length tTG in astrocyte cells treated with the inflammatory cytokines IL-1β and TNF-α (16), in rat brain injury and cerebral artery occlusion (35, 36), and in spinal cord injury models (37). However, to what extent this form was derived from leukocyte infiltration cannot be determined. These data suggest that the induction of tTGV1 and tTGV2 may be associated with the body’s response to injury. Like tTGV1 and tTGV2, the spliced site of rat S-TGN also is located at amino acid 622 and generates an alternate 30 amino acids at the C-terminal end; it was shown to be less sensitive to GTP-mediated inhibition of TGase activity (16, 34). However, the specific activity of rat s-TGN with respect to full-length tTG was not reported (16, 34). In addition, semiquantitative RT-PCR assays were used in these studies (35-37) and amore accurate measurement of the relative transcript levels is needed.
The cellular expression of tTG is considered ubiquitous (6, 8), but the quantitative expression comparing different cell types at the mRNA level has not been previously reported (Table 1). Our data are consistent with other reports that HUVECs and VSMCs are abundant in tTG (38). In addition to the dramatic differences in expression of tTG and tTGV1/tTGV2 mRNAs between HUVECs and leukocytes, the mRNA ratios of tTG/tTGV1 and tTG/tTGV2 also showed significant differences (Table 1). In HUVECs, tTG mRNA was expressed at 32- and 194-fold higher levels than tTGV1 and tTGV2, respectively, while in HL-60 and leukocytes (MN and PMN cells), the difference between tTG mRNA and tTGV1 (tTGV2) mRNA was much smaller (Table 1). RA-treatment of HL-60 cells dramatically increased the expression of tTG mRNA levels with ratios (tTG/tTGV1 and tTG/tTGV2) that were similar to HUVECs (Table 1). Immunoblot detection of tTG for HUVECs was consistent with the mRNA expression (Fig. 2). However, tTGV1 was expressed predominantly in leukocytes enriched in MNs, although there was more tTG mRNA in MNs (Fig. 2). Elucidating the mechanism(s) underlying the stabilization of tTGV1 in MNs requires further investigation. The induction of tTG on RA-treatment in HL-60 is associated with leukemia cell differentiation into macrophage-like cells (39). These data suggest that tTG plays a role in modulating cell differentiation, a conclusion supported by recent data showing that induction of tTG is important in regulating human neutrophils (PMNs) differentiation (39). However, a tTG-related antigen (similar to tTGV1 in mobility) also was detected in their immunoblots (Fig. 4b in their study) but the contribution was not discussed (39). The role of tTGV1/tTGV2 in this process warrants further investigation.
The differences in the mRNA expression among tTG, tTGV1, and tTGV2 in different cell types could be related to factor(s) that control tissue-specific transcription and/or mRNA stability and degradation. Methylation of the gene promoter is one of the mechanisms that control tissue-specific expression. The human tTG gene promoter was shown to be hypermethylated in HL-60 cells, monocytes, and lymphocytes, but not in HUVECs, and the methylation status is paralleled by low tTG expression and TGase activity levels in these cells (40, 41). However, RA is able to induce tTG expression in HL-60 cells that is independent of the hypermethylated tTG gene promoter, suggesting that RA regulates gene expression through a different mechanism (40). RA or inflammatory cytokines may induce proteins that can either stabilize or destabilize mRNAs, thereby changing their levels; this warrants further investigation (42, 43). At the protein level, we demonstrated that the in vivo stability of tTG, tTGV1, and tTGV2 can also be regulated due to their ability to bind GTP (Fig. 4). Our results indicated that the altered GTP binding ability of tTGV1/tTGV2 was the reason why they were poorly expressed in HEK293 cells. This is consistent with the observation that S171E/tTG, a point mutation GTP binding-deficient mutant, was poorly expressed and why there is a lack of TGase activity in NIH3T3 cells (14, 44). Another GTP-deficient mutant, rat R579A/tTG, also had relatively lower expression levels than wild-type tTG in SH-SY5Y cells (45). Its GTP-bound conformation may exclude tTG from the intracellular degradation pathway. This also warrants further investigation.
The unique localization of tTGV1 and tTGV2 in the cytoplasm of HUVECs and the nuclei of VSMCs suggests a specific intracellular function. One full-length 80 kDa and a 36 kDa truncated form of tTG have been detected in rabbit liver nuclei (46). tTG also has been found in the nuclei of Huntington’s Disease brain cells (47) and was recently reported to translocate to the nucleus in leukemia cells undergoing differentiation following RA treatment (39). It is still unclear at present how proteins are imported into the nucleus; the interaction of tTG with importin-α3 and/or phospholipase Cδ1 (PLCδ1) may be one such mechanism (6, 48, 49; see Discussion below).
X-ray crystallography reveals that tTG is composed of an N-terminal β-sandwich (domain I; residue #1-139), a α/β catalytic core (domain II; residue #140-454), a β-barrel 1 (domain III; residue #479-585), and a β-barrel 2 (Domain IV; residue #586-687) (27). The TGase active site is composed of a catalytic triad of C277-H335-D358 (27), and the rate-limiting step in catalysis involves the formation of a transitional thioester bond between C277 and the Q substrate (6, 8). The loss of part of domain IV (C-terminal 55 amino acids) dramatically reduced Ca+2 dependent TGase activity, since tTGV1 and tTGV2 had only 8-7% residual activity (Table 2). These data are consistent with our previous report that the C-terminal truncated form of tTG, M1-S538 (the ΔS538 mutant is similar in length to tTGH with part of domain III and all of domain IV deleted), has <5% of TGase activity (18). Recent X-ray crystallography and mutagenesis studies demonstrated a coordinated interaction between domains II to IV to achieve substrate specificity and efficient catalysis (50-53). Catalysis requires Ca+2 binding, the widening of the active site pocket, and interaction with the Q-substrate to form a thioester bond (6, 51, 52). Molecular modeling indicated that H658, K672, L673, K674, and A675 (deleted in tTGV1 and tTGV2) are involved in the interaction with D25, R28 in domain I, M485 in domain III, and R552 and E557 in domain IV (pfam website in Materials and Methods; ref. 27). Therefore, the C-terminal residues of tTG may be important in maintaining the overall tertiary structure of the protein. These data demonstrated that a complete tTG structure with four domains is necessary to achieve optimum catalysis (18).
tTGV1 and tTGV2 were unable to adopt a protease-resistant GTP-bound conformation and were insensitive to GTP-mediated inhibition of TGase activity. Bio-physical data including small-angle neutron, circular dichroism, and X-ray scattering demonstrate that there is a slight widening of tTG’s structure on Ca+2 binding, whereas there is a more compact structure in the presence of GTP (50-52). R580 was shown to be an important residue in GTP binding and involved in transition to a compact inactive TGase form of tTG by forming a stabilizing H-bond between C277-Y516 (53). Based on the 3-D structure of GDP-bound tTG (pdb: 1kv3), GTP binding involves the amino acid side chains from domains II, III, and IV binding to hydrophobic guanine binding pocket (K173, F174, S482, Met483, L582, and Y583), α, β-phosphates (R478, V479, R580, and Y583), and γ-phosphate (K173 and R476) (27). R476, R478, V479, S482, and M483 are located at the beginning of domain III, sitting within a β-hairpin region that involves two β-strand peptide fragments, M473-R478 and F489-N497 connected by a β-turn V479-D488 peptide (27). The M485 and G486 within this β-turn are involved in H-bonding with P612, L613, P614, and L673 in domain IV (27). The deletion of the AAs after 622 in tTGV1 and tTGV2 could alter the interaction with GTP, which would prevent the protein from adopting a protease-resistant conformation. This possibility is supported by data showing that the TGase activity of tTGV1 and tTGV2 was not inhibited by GTP (up to 400 μM) and remained sensitive to trypsin degradation (Fig. 3) in the presence of GTP.
The GTP binding domain was further investigated using a fluorescent GTP analog, BODIPY FL-GTP (Fig. 4). The mechanism for the increase in fluorescence is due to the movement of the fluorescent BODIPY moiety away from the guanine base on binding to the guanine binding pocket of the G-protein, thus relieving quenching between BODIPY and guanine and resulting in an increased fluorescence quantum yield (23, 54). A BODIPY FL-GTP binding assay demonstrated that the guanine binding pocket was altered in tTGV1/tTGV2, as there was little increase in fluorescence on binding (Fig. 4). However, the binding to the hydrolysis domain involving phosphate groups appeared to be unaltered because tTGV1 and tTGV2 were able to carry out Mg+2-dependent GTP hydrolysis (Fig. 4). The mechanism of the 6- to 7-fold increase in GTP hydrolysis might be due to more efficient access to the phosphate groups for hydrolysis and is consistent with our previous findings that C-terminal truncation mutant of tTG, ΔS538, exhibited enhanced GTP hydrolysis activity (18). The GTPase activity of tTG is important in cell-cycle arrest at the S to G2/M interphase and is linked to the α1-adrenergic-mediated signaling pathway (3, 55). In this respect, the significance of the altered expression of tTGV1/tTGV2 in cell differentiation and G-protein signaling remains to be determined. We also do not know whether the kinase activity of tTGV1/tTGV2 is altered. The kinase activity of tTG has been reported to phosphorylate histones in the nuclei of breast cancer cells (4, 5). This suggests that the kinase activity of tTG and its isoforms could play a role in controlling chromatin structure or in controlling other kinase-mediated signaling events.
Under normal conditions, intracellular free Ca+2 (∼10-7 M) and GTP (∼100-150 μM) are sufficient to keep the TGase activity of tTG in a latent state and prevent cross-linking of intracellular proteins (9). Since tTGV1 and tTGV2 are not readily inhibited by GTP, they could be readily activated by any transient intracellular Ca+2 spike stimulated by oxidative stress (56), which has been observed in many neurodegenerative diseases with mitochondria dysfunction (57, 58) or under other conditions. Although tTGV1,2 have less TGase activity, the constitutive crosslinking activity could allow for the gradual accumulation of cross-linked products over a long time period, which could result in further injury to the brain and explain why such diseases manifest themselves so late in life. Alternatively, local intracellular concentrations may provide sufficient levels of Ca+2 to promote local and specific tTG crosslinking. TGase activity of tTG was recently reported to function as a cofactor to cross-link Ikbα and activate NF-κB in the nuclei of brain astrocytes and BV-2 microglia (59, 60) (61). This raises the possibility that tTGV1 and tTGV2 might play a role in the NFkappaB activation process and can be easily activated by Ca+2.
The loss of C-terminal residues in the splice variants may affect their ability to form complexes with other proteins. Recently, tTG secreted in the ECM was found to serve as a ligand for GPR56, an orphan G-protein coupled receptor that is involved in suppressing tumor growth and metastasis (11). The N-terminal domain of GPR56 was found to bind to the C-terminal β-barrels of tTG (11). Since the C-terminal β-barrels are required for tTG to bind GPR56 (11), tTGV1 and tTGV2 may no longer serve as adhesive ligands for GPR56. Studies are in progress to test this hypothesis. In addition, the C-terminal AAs (#661-672) of tTG are involved in PLCδ1 binding (49). The activity of PLCδ1 is suppressed by the interaction with tTG (62). When GTP binds to tTG, PLCδ1 is released from inhibition (62). PLCδ1 shuttles between the cytoplasm and the nucleus (63, 64). By lacking critical C-terminal amino acids, tTGV1 and tTGV2 may have a defect in PLCδ1 binding and be unable to adopt a GTP-bound conformation, suggesting that PLCδ1 function, including its nuclear activity, might be enhanced. The interaction of tTG with phospholipase Cδ1 (PLCδ1) and/or importin-α3 may be an important mechanism that regulates its distribution between cytosol and nucleus (6, 48, 49). Thus, the potential loss of interaction with PLCδ1 may contribute to the unique pattern of intracellular localization of the splice variants. The N-terminal residues of tTG are preserved in the splice variants. tTG binds FN with high affinity through its N-terminal β-sandwich domain, aiding in the nonclassical secretion and localization of tTG on the cell surface (6, 65, 66). tTGV1/V2 were found to bind FN to a similar extent as tTG suggesting they can also be secreted (Fig. 3D).
As HEK293 cells express nondetectable tTG-related antigen, it provides a good cellular model to study the role of different forms of tTG. HEK293 expressing tTG, tTGV1, and tTGV2 showed little difference in LDH release, cell viability, and caspases 3/7 activation when compared with vector transfected cells (see Supplemental Fig. 5). The caspase activation is also consistent with the immunoblot data that PARP (in vivo substrate of caspases) cleavage was not detected (Fig, 4). These data suggest that the expression of tTG, tTGV1,or tTGV2 did not contribute to cell necrosis and apoptosis. When this paper was prepared for submission, Antonyak et al. reported the cellular function of two C-terminal truncated forms of tTG lacking C-terminal 139 (designated TGase-S; same as tTGH, tTG-S) and 30 amino acid [designated TGase (1-657); ref. 33]. In contrast to the protective role of tTG, tTG-S and TGase (1-657) were found to be cytotoxic to the cells and the effects were independent of their TGase activity (33). Aberrant high molecular mass oligomeric aggregates of TGase-S were found in dying cells (33). Their expression constructs had several notable differences from ours that might explain why we did not observe cell toxicity or high m aggregates in HEK293 cells transfected with tTGV1/tTGV2. These differences include the presence of fusion myc-tag (10 AAs) residues (33) and the inclusion of alternate 52 and 23 AAs after AA 622 at the C terminus of tTGV1 and tTGV2 (Fig. 1C).
In summary, tTGV1 and tTGV2 were abundantly expressed in primary HUVEC and VSMC, and TTGV1 was detected as a major tTG antigen in human leukocytes, suggesting that these isoforms are necessary to maintain normal cellular function. Indeed, tTGV1 is the major species in leukocytes suggesting important roles for it in these differentiated cells. The detection of alternative splicing of tTG provides a molecular explanation for our prior observation of lower m forms of tTG antigens in HUVECs and VSMCs. Further studies will be needed to ascertain whether tTGV1 and tTGV2 can modulate the multitude of biological functions ascribed to tTG including its roles in vascular biology, neurodegenerative diseases, inflammation and wound healing.
Supplementary Material
Acknowledgments
T.-S. Lai and C. S. Greenberg initiated design of the experiments, writing and revising the manuscript. T.-S. Lai, W. Li, and Y. Liu performed experiments and prepared the figures. This research was funded in part by NIH grants HL072184 (C. S. Greenberg) and NS050541 (T.-S. Lai), a Department of Defense award (C. S. Greenberg), a grant-in-aid from the American Heart Association (T.-S. Lai), support from the Huntington Disease Society of America (HDSA) (T.-S. Lai), and a Donald A. King Student Fellowship from HDSA (Y. Liu). The excellent technical assistance of Pei Yen and Vanessa Dorismond is greatly appreciated. The authors would like to thank Dr. David C. Sane at Wake Forest School of Medicine for critical reading of the manuscript and the Duke microarray facility for the initial real-time TaqMan assays.
REFERENCES
- 1.Folk JE. Mechanism and basis for specificity of transglutaminase-catalyzed epsilon-(gamma-glutamyl) lysine bond formation. Adv. Enzymol. Relat. Areas. Mol. Biol. 1983;54:1–56. doi: 10.1002/9780470122990.ch1. [DOI] [PubMed] [Google Scholar]
- 2.Achyuthan KE, Greenberg CS. Identification of a guanosine triphosphate-binding site on guinea pig liver transglutaminase. Role of GTP and calcium ions in modulating activity. J. Biol. Chem. 1987;262:1901–1906. [PubMed] [Google Scholar]
- 3.Nakaoka H, Perez DM, Baek KJ, Das T, Husain A, Misono K, Im MJ, Graham RM. Gh: a GTP-binding protein with transglutaminase activity and receptor signaling function. Science. 1994;264:1593–1596. doi: 10.1126/science.7911253. [DOI] [PubMed] [Google Scholar]
- 4.Mishra S, Murphy LJ. Tissue transglutaminase has intrinsic kinase activity: identification of transglutaminase 2 as an insulin-like growth factor-binding protein-3 kinase. J. Biol. Chem. 2004;279:23863–23868. doi: 10.1074/jbc.M311919200. [DOI] [PubMed] [Google Scholar]
- 5.Mishra S, Saleh A, Espino PS, Davie JR, Murphy LJ. Phosphorylation of histones by tissue transglutaminase. J. Biol. Chem. 2006 doi: 10.1074/jbc.M506864200. [DOI] [PubMed] [Google Scholar]
- 6.Lorand L, Graham RM. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 7.Iismaa SE, Begg GE, Graham RM. Cross-linking transglutaminases with G protein-coupled receptor signaling. Sci. STKE. 20062006:pe34. doi: 10.1126/stke.3532006pe34. [DOI] [PubMed] [Google Scholar]
- 8.Greenberg CS, Birckbichler PJ, Rice RH. Transglutaminases: multifunctional cross-linking enzymes that stabilize tissues. FASEB J. 1991;5:3071–3077. doi: 10.1096/fasebj.5.15.1683845. [DOI] [PubMed] [Google Scholar]
- 9.Lai TS, Slaughter TF, Peoples KA, Hettasch JM, Greenberg CS. Regulation of human tissue transglutaminase function by magnesium-nucleotide complexes. Identification of distinct binding sites for Mg-GTP and Mg-ATP. J. Biol. Chem. 1998;273:1776–1781. doi: 10.1074/jbc.273.3.1776. [DOI] [PubMed] [Google Scholar]
- 10.Lai T, Tucker T, Burke JR, Strittmatter WJ, Greenberg CS. Effect of tissue transglutaminase on the solubility of proteins containing expanded polyglutamine repeats. J. Neurochemistry. 2004;88:1253–1260. doi: 10.1046/j.1471-4159.2003.02249.x. [DOI] [PubMed] [Google Scholar]
- 11.Xu L, Begum S, Hearn JD, Hynes RO. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9023–9028. doi: 10.1073/pnas.0602681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sane DC, Kontos JL, Greenberg CS. Roles of transglutaminases in cardiac and vascular diseases. Front. Biosci. 2007;12:2530–2545. doi: 10.2741/2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonyak MA, Miller AM, Jansen JM, Boehm JE, Balkman CE, Wakshlag JJ, Page RL, Cerione RA. Augmentation of tissue transglutaminase expression and activation by epidermal growth factor inhibit doxorubicin-induced apoptosis in human breast cancer cells. J. Biol. Chem. 2004;279:41461–41467. doi: 10.1074/jbc.M404976200. [DOI] [PubMed] [Google Scholar]
- 14.Antonyak MA, Singh US, Lee DA, Boehm JE, Combs C, Zgola MM, Page RL, Cerione RA. Effects of tissue transglutaminase on retinoic acid-induced cellular differentiation and protection against apoptosis. J. Biol. Chem. 2001;276:33582–33587. doi: 10.1074/jbc.M105318200. [DOI] [PubMed] [Google Scholar]
- 15.Fraij BM, Gonzales RA. Organization and structure of the human tissue transglutaminase gene. Biochim. Biophys. Acta. 1997;1354:65–71. doi: 10.1016/s0167-4781(97)00132-2. [DOI] [PubMed] [Google Scholar]
- 16.Monsonego A, Shani Y, Friedmann I, Paas Y, Eizenberg O, Schwartz M. Expression of GTP-dependent and GTP-independent tissue-type transglutaminase in cytokine-treated rat brain astrocytes. J. Biol. Chem. 1997;272:3724–3732. doi: 10.1074/jbc.272.6.3724. [DOI] [PubMed] [Google Scholar]
- 17.Haroon ZA, Wannenburg T, Gupta M, Greenberg CS, Wallin R, Sane DC. Localization of tissue transglutaminase in human carotid and coronary artery atherosclerosis: implications for plaque stability and progression. Lab. Invest. 2001;81:83–93. doi: 10.1038/labinvest.3780214. [DOI] [PubMed] [Google Scholar]
- 18.Lai TS, Slaughter TF, Koropchak CM, Haroon ZA, Greenberg CS. C-terminal deletion of human tissue transglutaminase enhances magnesium-dependent GTP/AT-Pase activity. J. Biol. Chem. 1996;271:31191–31195. doi: 10.1074/jbc.271.49.31191. [DOI] [PubMed] [Google Scholar]
- 19.Citron BA, SantaCruz KS, Davies PJ, Festoff BW. Intron-exon swapping of transglutaminase mRNA and neuronal Tau aggregation in Alzheimer’s disease. J. Biol. Chem. 2001;276:3295–3301. doi: 10.1074/jbc.M004776200. [DOI] [PubMed] [Google Scholar]
- 20.Lesort M, Attanavanich K, Zhang J, Johnson GV. Distinct nuclear localization and activity of tissue transglutaminase. J. Biol. Chem. 1998;273:11991–11994. doi: 10.1074/jbc.273.20.11991. [DOI] [PubMed] [Google Scholar]
- 21.Boyum A. Separation of white blood cells. Nature. 1964;204:793–794. doi: 10.1038/204793a0. [DOI] [PubMed] [Google Scholar]
- 22.Slaughter TF, Achyuthan KE, Lai TS, Greenberg CS. A microtiter plate transglutaminase assay utilizing 5-(biotinamido)pentylamine as substrate. Anal. Biochem. 1992;205:166–171. doi: 10.1016/0003-2697(92)90594-w. [DOI] [PubMed] [Google Scholar]
- 23.McEwen DP, Gee KR, Kang HC, Neubig RR. Fluorescence approaches to study G protein mechanisms. Methods Enzymol. 2002;344:403–420. doi: 10.1016/s0076-6879(02)44730-1. [DOI] [PubMed] [Google Scholar]
- 24.Datta S, Antonyak MA, Cerione RA. Importance of Ca(2+)-Dependent transamidation activity in the protection afforded by tissue transglutaminase against doxorubicin-induced apoptosis. Biochemistry. 2006;45:13163–13174. doi: 10.1021/bi0606795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Achyuthan KE, Rowland TC, Birckbichler PJ, Lee KN, Bishop PD, Achyuthan AM. Hierarchies in the binding of human factor XIII, factor XIIIa, and endothelial cell transglutaminase to human plasma fibrinogen, fibrin, and fibronectin. Mol. Cell Biochem. 1996;162:43–49. doi: 10.1007/BF00250994. [DOI] [PubMed] [Google Scholar]
- 26.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′—-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S, Cerione RA, Clardy J. Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2743–2747. doi: 10.1073/pnas.042454899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraij BM, Gonzales RA. A third human tissue transglutaminase homologue as a result of alternative gene transcripts. Biochim. Biophys. Acta. 1996;1306:63–74. doi: 10.1016/0167-4781(95)00219-7. [DOI] [PubMed] [Google Scholar]
- 29.Willard FS, Kimple AJ, Johnston CA, Siderovski DP. A direct fluorescence-based assay for RGS domain GT-Pase accelerating activity. Anal. Biochem. 2005;340:341–351. doi: 10.1016/j.ab.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Boehm JE, Singh U, Combs C, Antonyak MA, Cerione RA. Tissue transglutaminase protects against apoptosis by modifying the tumor suppressor protein p110 Rb. J. Biol. Chem. 2002;277:20127–20130. doi: 10.1074/jbc.C200147200. [DOI] [PubMed] [Google Scholar]
- 31.Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 32.Citron BA, Suo Z, SantaCruz K, Davies PJ, Qin F, Festoff BW. Protein crosslinking, tissue transglutaminase, alternative splicing and neurodegeneration. Neurochem. Int. 2002;40:69–78. doi: 10.1016/s0197-0186(01)00062-6. [DOI] [PubMed] [Google Scholar]
- 33.Antonyak MA, Jansen JM, Miller AM, Ly TK, Endo M, Cerione RA. Two isoforms of tissue transglutaminase mediate opposing cellular fates. Proc. Natl. Acad. Sci. U. S. A. 2006 doi: 10.1073/pnas.0604844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monsonego A, Friedmann I, Shani Y, Eisenstein M, Schwartz M. GTP-dependent conformational changes associated with the functional switch between Galpha and cross-linking activities in brain-derived tissue transglutaminase. J. Mol. Biol. 1998;282:713–720. doi: 10.1006/jmbi.1998.2052. [DOI] [PubMed] [Google Scholar]
- 35.Tolentino PJ, DeFord SM, Notterpek L, Glenn CC, Pike BR, Wang KK, Hayes RL. Up-regulation of tissue-type transglutaminase after traumatic brain injury. J. Neurochem. 2002;80:579–588. doi: 10.1046/j.0022-3042.2001.00726.x. [DOI] [PubMed] [Google Scholar]
- 36.Tolentino PJ, Waghray A, Wang KK, Hayes RL. Increased expression of tissue-type transglutaminase following middle cerebral artery occlusion in rats. J. Neurochem. 2004;89:1301–1307. doi: 10.1111/j.1471-4159.2004.02436.x. [DOI] [PubMed] [Google Scholar]
- 37.Festoff BW, SantaCruz K, Arnold PM, Sebastian CT, Davies PJ, Citron BA. Injury-induced “switch” from GTP-regulated to novel GTP-independent isoform of tissue transglutaminase in the rat spinal cord. J. Neurochem. 2002;81:708–718. doi: 10.1046/j.1471-4159.2002.00850.x. [DOI] [PubMed] [Google Scholar]
- 38.Thomazy V, Fesus L. Differential expression of tissue transglutaminase in human cells. An immunohistochemical study. Cell Tissue Res. 1989;255:215–224. doi: 10.1007/BF00229084. [DOI] [PubMed] [Google Scholar]
- 39.Balajthy Z, Csomos K, Vamosi G, Szanto A, Lanotte M, Fesus L. Tissue-transglutaminase contributes to neutrophil granulocyte differentiation and functions. Blood. 2006 doi: 10.1182/blood-2004-02-007948. [DOI] [PubMed] [Google Scholar]
- 40.Lu S, Davies PJ. Regulation of the expression of the tissue transglutaminase gene by DNA methylation. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4692–4697. doi: 10.1073/pnas.94.9.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cacciamani T, Virgili S, Centurelli M, Bertoli E, Eremenko T, Volpe P. Specific methylation of the CpG-rich domains in the promoter of the human tissue transglutaminase gene. Gene. 2002;297:103–112. doi: 10.1016/s0378-1119(02)00874-0. [DOI] [PubMed] [Google Scholar]
- 42.Otake Y, Sengupta TK, Bandyopadhyay S, Spicer EK, Fernandes DJ. Drug-induced destabilization of bcl-2 mRNA: a new approach for inducing apoptosis in tumor cells. Curr. Opin. Investig. Drugs. 2004;5:616–622. [PubMed] [Google Scholar]
- 43.Mueller WH, Kleefeld D, Khattab B, Meissner JD, Scheibe RJ. Effects of retinoic acid on N-glycosylation and mRNA stability of the liver/bone/kidney alkaline phosphatase in neuronal cells. J. Cell Physiol. 2000;182:50–61. doi: 10.1002/(SICI)1097-4652(200001)182:1<50::AID-JCP6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 44.Jeon JH, Cho SY, Kim CW, Shin DM, Kweon JC, Choi KH, Park SC, Kim IG. GTP is required to stabilize and display transamidation activity of transglutaminase 2. Biochem. Biophys. Res. Commun. 2002;294:818–822. doi: 10.1016/S0006-291X(02)00582-X. [DOI] [PubMed] [Google Scholar]
- 45.Begg GE, Holman SR, Stokes PH, Matthews JM, Graham RM, Iismaa SE. Mutation of a critical GTP-binding arginine in transglutaminase 2 disinhibits intracellular crosslinking activity. J. Biol. Chem. 2006 doi: 10.1074/jbc.M600146200. [DOI] [PubMed] [Google Scholar]
- 46.Singh US, Erickson JW, Cerione RA. Identification and biochemical characterization of an 80 kilo-dalton GTP-binding/transglutaminase from rabbit liver nuclei. Biochemistry. 1995;34:15863–15871. doi: 10.1021/bi00048a032. [DOI] [PubMed] [Google Scholar]
- 47.Karpuj MV, Garren H, Slunt H, Price DL, Gusella J, Becher MW, Steinman L. Transglutaminase aggregates huntingtin into nonamyloidogenic polymers, and its enzymatic activity increases in Huntington’s disease brain nuclei. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7388–7393. doi: 10.1073/pnas.96.13.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng X, Zhang Y, Zhang H, Graner S, Williams JF, Levitt ML, Lokshin A. Interaction of tissue transglutaminase with nuclear transport protein importin-alpha3. FEBS Lett. 1999;446:35–39. doi: 10.1016/s0014-5793(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 49.Im MJ, Russell MA, Feng JF. Transglutaminase II: a new class of GTP-binding protein with new biological functions. Cell Signal. 1997;9:477–482. doi: 10.1016/s0898-6568(97)00049-1. [DOI] [PubMed] [Google Scholar]
- 50.Casadio R, Polverini E, Mariani P, Spinozzi F, Carsughi F, Fontana A, Polverino de Laureto P, Matteucci G, Bergamini CM. The structural basis for the regulation of tissue transglutaminase by calcium ions. Eur. J. Biochem. 1999;262:672–679. doi: 10.1046/j.1432-1327.1999.00437.x. [DOI] [PubMed] [Google Scholar]
- 51.Mariani P, Carsughi F, Spinozzi F, Romanzetti S, Meier G, Casadio R, Bergamini CM. Ligand-induced conformational changes in tissue transglutaminase: Monte Carlo analysis of small-angle scattering data. Biophys. J. 2000;78:3240–3251. doi: 10.1016/S0006-3495(00)76860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Venere A, Rossi A, De Matteis F, Rosato N, Agro AF, Mei G. Opposite effects of Ca(2+) and GTP binding on tissue transglutaminase tertiary structure. J. Biol. Chem. 2000;275:3915–3921. doi: 10.1074/jbc.275.6.3915. [DOI] [PubMed] [Google Scholar]
- 53.Begg GE, Carrington L, Stokes PH, Matthews JM, Wouters MA, Husain A, Lorand L, Iismaa SE, Graham RM. Mechanism of allosteric regulation of transglutaminase 2 by GTP. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19683–19688. doi: 10.1073/pnas.0609283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McEwen DP, Gee KR, Kang HC, Neubig RR. Fluorescent BODIPY-GTP analogs: real-time measurement of nucleotide binding to G proteins. Anal. Biochem. 2001;291:109–117. doi: 10.1006/abio.2001.5011. [DOI] [PubMed] [Google Scholar]
- 55.Mian S, el Alaoui S, Lawry J, Gentile V, Davies PJ, Griffin M. The importance of the GTP-binding protein tissue transglutaminase in the regulation of cell cycle progression. FEBS Lett. 1995;370:27–31. doi: 10.1016/0014-5793(95)00782-5. [DOI] [PubMed] [Google Scholar]
- 56.Shin DM, Jeon JH, Kim CW, Cho SY, Kwon JC, Lee HJ, Choi KH, Park SC, Kim IG. Cell type-specific activation of intracellular transglutaminase 2 by oxidative stress or ultraviolet irradiation: implications of transglutaminase 2 in age-related cataractogenesis. J. Biol. Chem. 2004;279:15032–15039. doi: 10.1074/jbc.M308734200. [DOI] [PubMed] [Google Scholar]
- 57.Fox JH, Barber DS, Singh B, Zucker B, Swindell MK, Norflus F, Buzescu R, Chopra R, Ferrante RJ, Kazantsev A, Hersch SM. Cystamine increases L-cysteine levels in Huntington’s disease transgenic mouse brain and in a PC12 model of polyglutamine aggregation. J. Neurochem. 2004;91:413–422. doi: 10.1111/j.1471-4159.2004.02726.x. [DOI] [PubMed] [Google Scholar]
- 58.Minghetti L. Role of inflammation in neurodegenerative diseases. Curr. Opin. Neurol. 2005;18:315–321. doi: 10.1097/01.wco.0000169752.54191.97. [DOI] [PubMed] [Google Scholar]
- 59.Lee J, Kim YS, Choi DH, Bang MS, Han TR, Joh TH, Kim SY. Transglutaminase 2 induces nuclear factor-kappaB activation via a novel pathway in BV-2 microglia. J. Biol. Chem. 2004;279:53725–53735. doi: 10.1074/jbc.M407627200. [DOI] [PubMed] [Google Scholar]
- 60.Caccamo D, Campisi A, Curro M, Aguennouz M, Li Volti G, Avola R, Ientile R. Nuclear factor-kappab activation is associated with glutamate-evoked tissue transglutaminase up-regulation in primary astrocyte cultures. J. Neurosci. Res. 2005;82:858–865. doi: 10.1002/jnr.20683. [DOI] [PubMed] [Google Scholar]
- 61.Mann AP, Verma A, Sethi G, Manavathi B, Wang H, Fok JY, Kunnumakkara AB, Kumar R, Aggarwal BB, Mehta K. Overexpression of tissue transglutaminase leads to constitutive activation of nuclear factor-{kappa}B in cancer cells: delineation of a novel pathway. Cancer Res. 2006;66:8788–8795. doi: 10.1158/0008-5472.CAN-06-1457. [DOI] [PubMed] [Google Scholar]
- 62.Murthy SN, Lomasney JW, Mak EC, Lorand L. Interactions of G(h)/transglutaminase with phospholipase Cdelta1 and with GTP. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11815–11819. doi: 10.1073/pnas.96.21.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamaga M, Fujii M, Kamata H, Hirata H, Yagisawa H. Phospholipase C-delta1 contains a functional nuclear export signal sequence. J. Biol. Chem. 1999;274:28537–28541. doi: 10.1074/jbc.274.40.28537. [DOI] [PubMed] [Google Scholar]
- 64.Irvine RF. 20 years of Ins(1,4,5)P3, and 40 years before. Nat. Rev. Mol. Cell Biol. 2003;4:586–590. doi: 10.1038/nrm1152. [DOI] [PubMed] [Google Scholar]
- 65.Hang J, Zemskov EA, Lorand L, Belkin AM. Identification of a novel recognition sequence for fibronectin within the NH2-terminal beta-sandwich domain of tissue transglutaminase. J. Biol. Chem. 2005;280:23675–23683. doi: 10.1074/jbc.M503323200. [DOI] [PubMed] [Google Scholar]
- 66.Akimov SS, Krylov D, Fleischman LF, Belkin AM. Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin. J. Cell Biol. 2000;148:825–838. doi: 10.1083/jcb.148.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.