Abstract
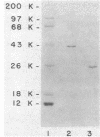
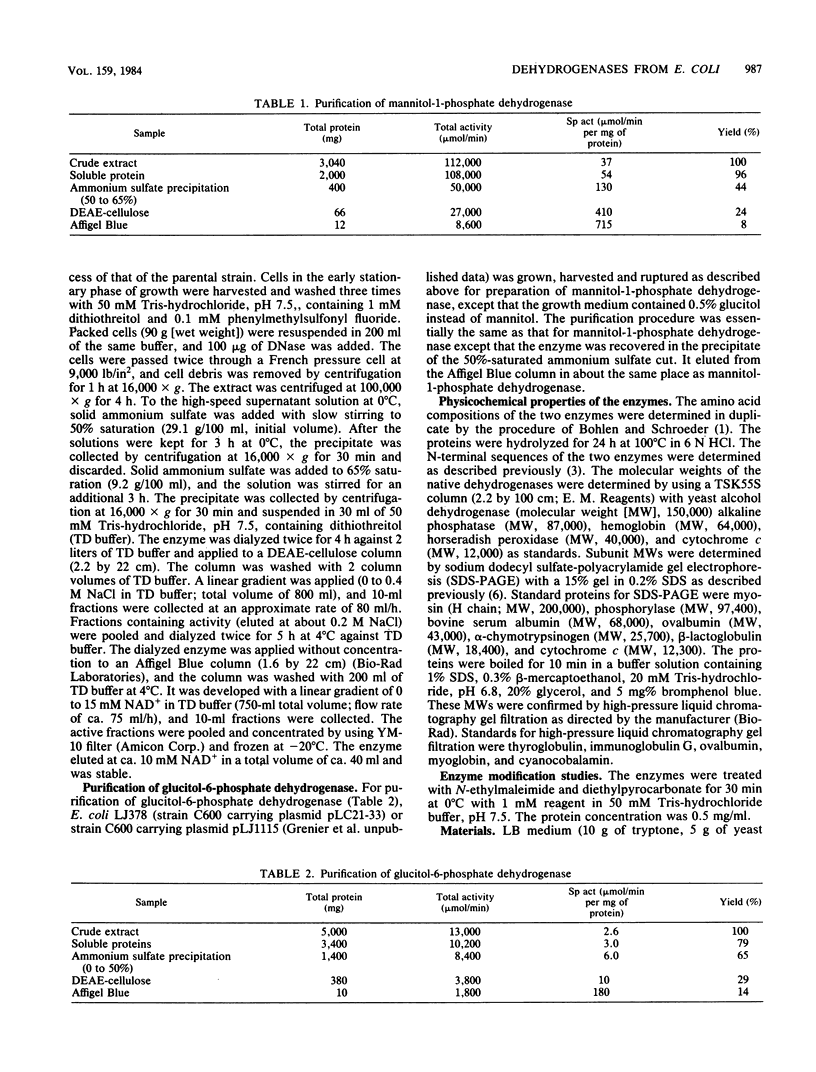
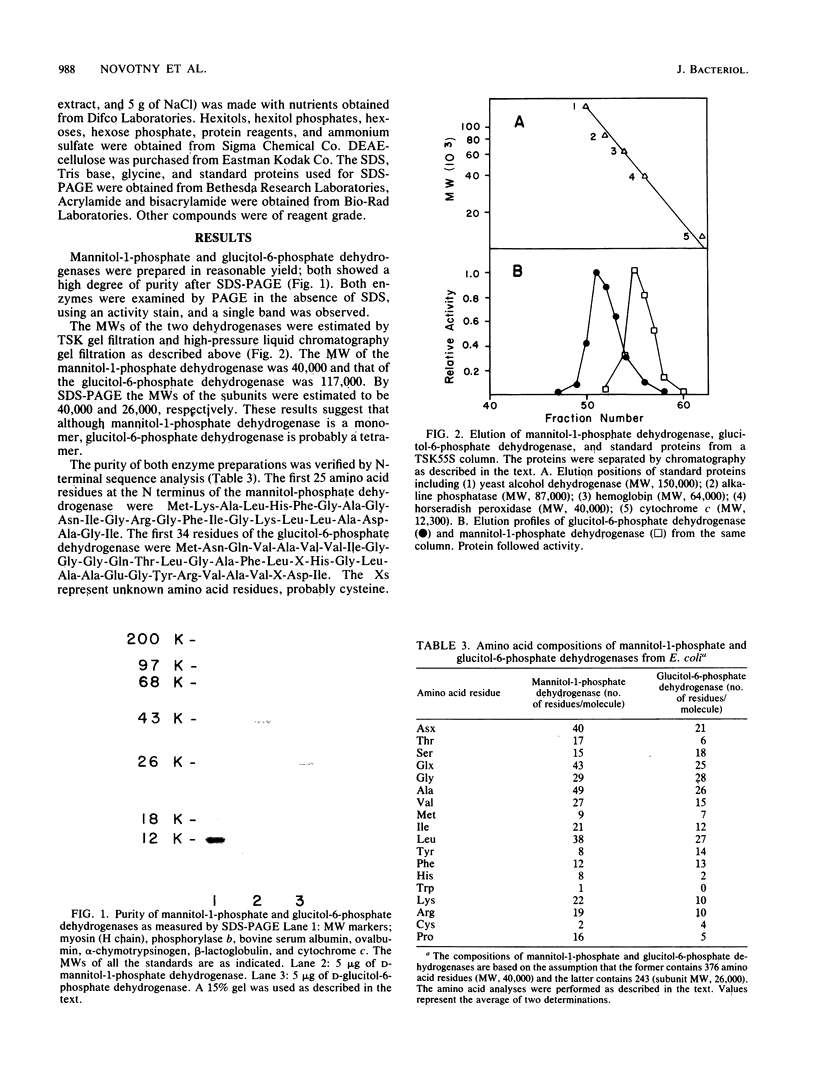
D-Mannitol-1-phosphate dehydrogenase (EC 1.1.1.17) and D-glucitol-6-phosphate dehydrogenase (EC 1.1.1.140) were purified to apparent homogeneity in good yields from Escherichia coli. The amino acid compositions, N-terminal amino acid sequences, sensitivities to chemical reagents, and catalytic properties of the two enzymes were determined. Both enzymes showed absolute specificities for their substrates. The subunit molecular weights of mannitol-1-phosphate and glucitol-6-phosphate dehydrogenases were 40,000 and 26,000, respectively; the apparent molecular weights of the native proteins, determined by gel filtration, were 40,000 and 117,000, respectively. It is therefore concluded that whereas mannitol-1-phosphate dehydrogenase is a monomer, glucitol-6-phosphate dehydrogenase is probably a tetramer. These two proteins differed in several fundamental respects.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dills S. S., Apperson A., Schmidt M. R., Saier M. H., Jr Carbohydrate transport in bacteria. Microbiol Rev. 1980 Sep;44(3):385–418. doi: 10.1128/mr.44.3.385-418.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch F. S. Polypeptide microsequence analysis with the commercially available gas-phase sequencer. Anal Biochem. 1984 Jan;136(1):39–47. doi: 10.1016/0003-2697(84)90305-1. [DOI] [PubMed] [Google Scholar]
- Jacobson G. R., Lee C. A., Leonard J. E., Saier M. H., Jr Mannitol-specific enzyme II of the bacterial phosphotransferase system. I. Properties of the purified permease. J Biol Chem. 1983 Sep 10;258(17):10748–10756. [PubMed] [Google Scholar]
- Jacobson G. R., Lee C. A., Saier M. H., Jr Purification of the mannitol-specific enzyme II of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. J Biol Chem. 1979 Jan 25;254(2):249–252. [PubMed] [Google Scholar]
- LISS M., HORWITZ S. B., KAPLAN N. O. D-Mannitol 1-phosphate dehydrogenase and D-sorbitol 6-phosphate dehydrogenase in Aerobacter aerogenes. J Biol Chem. 1962 Apr;237:1342–1350. [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Lee C. A., Jacobson G. R., Saier M. H., Jr Plasmid-directed synthesis of enzymes required for D-mannitol transport and utilization in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7336–7340. doi: 10.1073/pnas.78.12.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. A., Saier M. H., Jr Mannitol-specific enzyme II of the bacterial phosphotransferase system. III. The nucleotide sequence of the permease gene. J Biol Chem. 1983 Sep 10;258(17):10761–10767. [PubMed] [Google Scholar]
- Lee C. A., Saier M. H., Jr Use of cloned mtl genes of Escherichia coli to introduce mtl deletion mutations into the chromosome. J Bacteriol. 1983 Feb;153(2):685–692. doi: 10.1128/jb.153.2.685-692.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler J. Analysis of mutations affecting the dissmilation of galactitol (dulcitol) in Escherichia coli K 12. Mol Gen Genet. 1977 Mar 28;152(1):83–91. doi: 10.1007/BF00264944. [DOI] [PubMed] [Google Scholar]
- Lengeler J. Mutations affecting transport of the hexitols D-mannitol, D-glucitol, and galactitol in Escherichia coli K-12: isolation and mapping. J Bacteriol. 1975 Oct;124(1):26–38. doi: 10.1128/jb.124.1.26-38.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler J. Nature and properties of hexitol transport systems in Escherichia coli. J Bacteriol. 1975 Oct;124(1):39–47. doi: 10.1128/jb.124.1.39-47.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler J., Steinberger H. Analysis of regulatory mechanisms controlling the activity of the hexitol transport systems in Escherichia coli K12. Mol Gen Genet. 1978 Nov 16;167(1):75–82. doi: 10.1007/BF00270323. [DOI] [PubMed] [Google Scholar]
- Leonard J. E., Saier M. H., Jr Genetic dissection of catalytic activities of the Salmonella typhimurium mannitol enzyme II. J Bacteriol. 1981 Feb;145(2):1106–1109. doi: 10.1128/jb.145.2.1106-1109.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard J. E., Saier M. H., Jr Mannitol-specific enzyme II of the bacterial phosphotransferase system. II. Reconstitution of vectorial transphosphorylation in phospholipid vesicles. J Biol Chem. 1983 Sep 10;258(17):10757–10760. [PubMed] [Google Scholar]
- Postma P. W., Roseman S. The bacterial phosphoenolpyruvate: sugar phosphotransferase system. Biochim Biophys Acta. 1976 Dec 14;457(3-4):213–257. doi: 10.1016/0304-4157(76)90001-0. [DOI] [PubMed] [Google Scholar]
- Sarno M. V., Tenn L. G., Desai A., Chin A. M., Grenier F. C., Saier M. H., Jr Genetic evidence for glucitol-specific enzyme III, an essential phosphocarrier protein of the Salmonella typhimurium glucitol phosphotransferase system. J Bacteriol. 1984 Mar;157(3):953–955. doi: 10.1128/jb.157.3.953-955.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni R. D., Roseman S., Saier M. H., Jr Sugar transport. Properties of mutant bacteria defective in proteins of the phosphoenolpyruvate: sugar phosphotransferase system. J Biol Chem. 1976 Nov 10;251(21):6584–6597. [PubMed] [Google Scholar]
- do Nascimento K. H., Davies D. D. The stereospecificity of sequential nicotinamide-adenine dinucleotide-dependent oxidoreductases in relation to the evolution of metabolic sequences. Biochem J. 1975 Sep;149(3):553–557. doi: 10.1042/bj1490553. [DOI] [PMC free article] [PubMed] [Google Scholar]