Abstract
The adult mammalian brain comprises many functionally distinct neuronal types, which are generated during development as a result of a coordinated signaling cascade that drives neuroblasts from proliferation into differentiation. We investigated whether and how ShcA adaptor proteins, which are known to function as initiators of the Ras signaling cascade in various nonneuronal systems where they have been considered to be expressed ubiquitously, are involved in the proliferative and differentiative phases of the developing brain. We found that in the forebrain expression and activation of ShcA proteins were strictly regulated during embryonic development, both temporally and spatially. The mRNAs encoded by the ShcA gene were expressed exclusively within an area to which active proliferation of immature neuroblasts was confined, the ventricular zone. In postnatal and adult brain, ShcA mRNAs and proteins were present only faintly. In the adult olfactory epithelium, in which neuronal cell renewal occurs throughout life, ShcA remained strongly expressed. These phenomena were peculiar to ShcA, since Grb2 adaptor protein remained expressed at constant level throughout development. The embryonically expressed ShcA proteins were functionally active, since p52ShcA became phosphorylated on tyrosine and associated with Grb2 following intraventricular injection of epidermal growth factor in the embryonic brain. Our data indicate that, through an orderly pattern of expression, ShcA gene products may play a role in the control of the switch between proliferation and differentiation of brain neuroblasts.
During mammalian neurogenesis, a temporally controlled and spatially localized availability of mitogenic and differentiative polypeptides and their corresponding receptors have been implicated in the series of events that ultimately lead proliferating neuroblasts to assume a terminally differentiated phenotype (1–3).
Many of the polypeptide growth factors known thus far exert their effects by interacting with cell surface receptors that contain tyrosine kinase (TK) domains and whose stimulation results in activation of the Ras-mitogen-activated protein kinase transduction pathway (4–6). The events upstream of Ras activation have recently been characterized for various growth factor receptors and have been shown to involve the recruitment of Grb2, a 23-kDa adaptor protein, and the guanine nucleotide release protein, Sos, by the activated receptors (7–10), ultimately leading to stimulation of the mitogen-activated protein kinase pathway (11, 12). Shc (from Src homologous and collagen) is another molecule involved in Ras activation (13). Indirect biochemical and functional evidence indicates that when a receptor such as that for epidermal growth factor (EGF) is activated, Shc adaptor protein rapidly binds to a specific phosphotyrosine on the stimulated receptor, becomes phosphorylated on Tyr-317 (13), and subsequently forms stable complexes with Grb2. Within this model of activation of the Ras pathway, following receptor stimulation, the constitutive complex Grb2–Sos is translocated from the cytosol to the membrane by interaction with the activated Shc, thereby leading to p21ras activation (14). Shc protein is also known to be involved in signaling from surface receptors devoid of intrinsic TK activity, probably through the recruitment of cytoplasmic TKs (15–19).
The Shc gene, recently renamed ShcA, encodes three proteins of 46 (p46shcA), 52 (p52shcA), and 66 (p66shcA) kDa, each containing an SH2–phosphotyrosine binding domain (20), a central glycine- and proline-rich region (collagen homologous region 1, CH1) (20), and a novel phosphotyrosine binding site named the PTB/PID domain (21–23). These domains are involved in multiple protein/protein interactions. Thus far, p46shcA and p52shcA isoforms, which originate from a different translation initiation site, were found to be expressed ubiquitously when considering cells of nonneuronal origin. These two proteins are indeed present and activated in fibroblasts, hepatocytes, and different immature and mature cells (i.e., T lymphocytes) of the hematopoietic system (20, 24–28). p66shcA protein is a less well-characterized molecule, expressed from a distinct transcript that is absent in some hematopoietic cells (20). Analyses of ShcA proteins functions have demonstrated their recruitment by various different activated receptors, including the EGF receptor (20, 24–28). ShcA proteins have oncogenic properties, because overexpression of p46shcA and p52shcA induces transformation in fibroblasts (20) and Ras-dependent differentiation in PC12 cells (29). Furthermore, in a recent report from Migliaccio et al. (30), p52/p46shcA proteins were found to activate transcription from the c-fos promoter. On the contrary, p66shcA is not capable of activating this same promoter and does not induce transformation in fibroblasts (30). The functional relevance of the ShcA gene is underscored by its conservation throughout vertebrate evolution (31). When considered together, these data indicate that ShcA proteins participate in a signaling pathway of fundamental significance for many mammalian cell types. Recently, two new genes have been identified that encode for two Shc proteins, termed ShcB/Sli and ShcC/Rai, which have been found to be particularly enriched in the adult mouse brain (32, 33). Although ShcA-driven effector systems have been well investigated in nonneuronal cells, a role during neurogenesis has not been considered thus far.
MATERIALS AND METHODS
Animals.
Sprague–Dawley pregnant rats of 14, 15, and 18 gestational days, neonates, and adult animals were obtained from Charles River Breeding Laboratories. Outbred CD-1 female mice (Charles River Breeding Laboratories) were used for the in situ analysis.
Primary Cultures.
Neuronal cultures were prepared from the embryonic day (E) 14 rat striatum primordia as described (34). Briefly, mechanically dissociated cells were plated onto polyornithine-coated 60-mm tissue culture dishes at a density of 3 × 105 cells per cm2. Cells were allowed to differentiate by growing them in serum-free medium (SFM) for different periods of time in vitro in a humidified incubator with 95% air/5% CO2. Cultured cells were rinsed three times in PBS and then lysed in lysis buffer (600 μl per plate). The collected material was handled as described below.
Antibodies.
Rabbit polyclonal antibody against ShcA isoforms was used at a ratio of 1:2,500 (Upstate Biotechnology, Lake Placid, NY). Anti-ShcA mAb derived from ascitic fluid (provided by P. G. Pelicci, Istituto Europeo di Oncologia, Milan) was applied at 1:3. mAb against Grb2 (Upstate Biotechnology) was applied at 1:1,000. mAb against α-tubulin (Sigma) was used at 1:5,000 dilution (this antibody recognizes all different isotypes of α-tubulin present in the rat brain). mAb against phosphotyrosines (Upstate Biotechnology) was applied at 1:1,000. Secondary antibodies (goat anti-rabbit or goat anti-mouse; Kirkegaard & Perry Laboratories) were used at 1:10,000 dilution.
Western Blot Analysis.
Lysates were obtained as described (35). Briefly, animals were sacrificed by cervical dislocation, the embryos/brains immediately collected, and the different brain regions (striatum, basal forebrain, cortex, hippocampus, and olfactory bulb) isolated using a dissecting microscope. The tissues were immediately frozen on dry ice. After weighing, tissues were homogenized using a teflon glass homogenizer in lysis buffer (10 μl/mg of tissue) (10% glycerol/50 mM Tris, pH 7.5/150 mM NaCl/5 mM EDTA/1 mM EGTA/1% Triton X-100/1 mM Na3VO4/1 mM ZnCl2 in the presence of 1 mM phenylmethylsulfonyl fluoride/10 mg/ml aprotinin/5 mg/ml leupeptin at 4°C). The extracts were passed several times through a 1-ml insulin syringe (26 gauge), incubated for 30 min at 4°C, and then cleared by centrifugation. Protein content was measured by a Bio-Rad protein assay kit. Aliquots of extracts were diluted with SDS sample buffer and boiled for 5 min. Equal amounts of proteins were loaded onto 10% SDS/PAGE. After transfer to nitrocellulose, the blots were blocked in 10% nonfat dry milk in TBS-T (20 mM Tris, pH 7.5/500 mM NaCl/0.01% Tween 20) overnight at 4°C. Blots were then incubated for 1 hr at room temperature in primary antibody. After washing with TBS-T, membranes were exposed to horseradish peroxidase-conjugated secondary antibody for 1 hr at room temperature. Immunoreactivities were detected using the enhanced chemiluminescence method (Amersham) according to the manufacturer’s instructions.
Immunoprecipitation.
Injection was performed following a procedure developed previously for embryonic transplantation of central nervous system cells (36). Ten microliters of EGF (50 ng/μl) was placed intraventricularly into E15 embryos. Control embryos were injected with vehicle. Ten minutes after injection, embryos were removed, the telencephalic vesicles dissected, lysed as previously described, and subjected to immunoprecipitation as described (35). ShcA polyclonal antibody (4 μg/sample) was used. The immunoprecipitated proteins were analyzed by Western blot analysis with the 4G10 anti-phosphotyrosine mAb or anti-Grb2 mAb.
Immunohistochemistry.
Pregnant females were injected intraperitoneally with BrdUrd (Boehringer Mannheim; 50 mg/kg dissolved in saline with 0.007 M NaOH) at gestational day 14. Injections were performed at intervals of 2 hr for a number of periods covering the whole cell cycle duration as estimated for that day of pregnancy. The dose of BrdUrd employed was previously found to saturate S-labeling for at least 2.0 hr after injection. Nuclei of the BrdUrd-containing cells were identified as described (36–39), and adjacent sections were used for in situ hybridization with Shc and p66ShcA probes. Injected animals were sacrificed by cervical dislocation and embryos dissected out in ice-cold PBS, fixed for 6 hr in 4% paraformaldehyde, and processed for paraffin embedding. Paraffin serial sections were cut at 7 μm on a microtome, mounted on gelatin-coated slides, and stored at 4°C, with silica gel, until use.
In Situ Analysis.
Embryos were collected from time-pregnant outbred CD-1 female mice (Charles River Breeding Laboratories) at E10.5–E12.5, fixed overnight in 4% paraformaldehyde in PBS and embedded in paraffin. Serial sections were cut at 7 μm on a microtome, mounted on gelatin coated slides, and stored at 4°C, with silica gel, until use. Brains from older animals were dissected out (E16.5, E18.5, and postnatal), fixed overnight in 4% paraformaldehyde in PBS, put in a 20% sucrose gradient overnight at 4°C, then in OCT embedding compound (Tissue-Tek), frozen in liquid nitrogen, and stored at −70°C until use. Serial sections (10 μm) were cut on a cryostat and mounted on gelatin-coated slides, washed in PBS twice, dehydrated in ethanol, and stored at −70°C until use. When different probes were compared, adjacent series were used. Two alternative series were used for each probe. Paraffin sections were dewaxed, rehydrated in ethanol, and processed for in situ hybridization as described by Wilkinson (40) and with minor modifications from Gulisano et al. (41). Cryostate sections were rehydrated and processed for in situ hybridization as described by Simmons et al. (42) and with minor modifications from Gulisano et al. (41). Autoradiography was performed with Kodak NT/B2 emulsion. Exposure times were between 5 and 12 days. After development, sections were counterstained with cresyl violet, mounted in DPX (Sigma), and photographed using either a stereomicroscope (Zeiss SV11) or an optical microscope (Zeiss Axiophot), with both dark- and bright-field illumination.
Shc and P66 sense and antisense [35S]-labeled RNA probes were synthetically produced using a 470-nt EcoRI–EcoRI fragment cloned in pGEM3 (Promega) (7) and a 370-nt EcoRI–EcoRI fragment cloned in pCRII (Invitrogen), respectively, as templates. Transcription reactions with T7 or SP6 polymerase (Riboprobe Kit, Promega) were carried out in the presence of [35S]CTP (Amersham) on linearized plasmids. The probes were resuspended at a working concentration of 1 × 105 cpm/μl in hybridization mix and stored at −80°C until use.
Densitometric Analysis.
The relative density of immunoreactive bands on Western blots was calculated from the area of the peak corresponding to the selected band following acquisition of the blot image through a Nikon CCD video camera module and analysis by means of the image 1.52 program (Wayne Rasband, National Institutes of Health, Research Services Branch, National Institute of Mental Health, Bethesda). Normalization over protein load was achieved by dividing each individual peak area by the corresponding peak area of the α-tubulin signal.
RESULTS
ShcA Expression During Neurogenesis in the Embryonic Murine Brain.
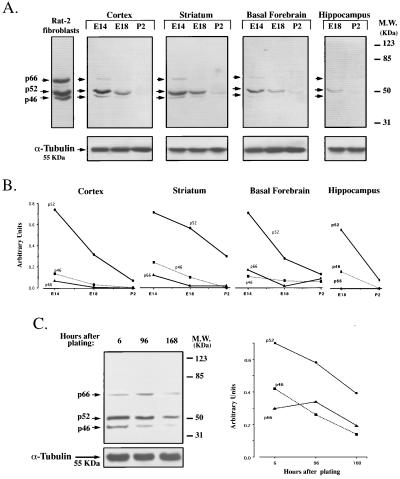
In an initial attempt to identify the intracellular mechanisms implicated in the responses to mitogenic and differentiative signals in neuroblasts, we analyzed whether ShcA proteins were expressed in the rodent forebrain at early stages of development. Western blot analyses, performed on lysates obtained from striatum, hippocampus, basal forebrain, and cortex revealed that p46shcA, p52shcA, and p66shcA were all present at high levels at the first gestational day analyzed (rat embryonic day 14, E14, for striatum, basal forebrain, and cortex; E18 for hippocampus) (Fig. 1A). At this embryonic stage, the majority of the cells are still immature and proliferating actively (43). When later developmental stages were considered (E18, P2) a sharp reduction in ShcA protein levels was observed in all of the brain regions analyzed. In Fig. 1B, the relative densities of the immunoreactive bands are plotted after normalization over an α-tubulin signal, used as an internal standard. Expression of all three ShcA isoforms decreases homogeneously during development. At all stages considered, the relative expression levels of the three isoforms were as follows: p52shcA > p46shcA > p66shcA. Identical results were obtained when analyzing tissues of mouse origin (data not shown).
Figure 1.
Expression of ShcA proteins during forebrain maturation. (A) Western blot analyses showing maximal expression of the three ShcA isoforms at early developmental stages in different brain regions. Equal amounts of protein (50 μg per lane) were electrophoresed. (Lower) Membranes were stripped and immunodecorated with anti-α-tubulin antibody. Lysates derived from Rat-2 fibroblasts were used as positive controls. E, embryonic; P, postnatal. (B) Densitometric analyses of p66ShcA, p52ShcA, and p46ShcA immunoreactive bands, normalized over α-tubulin content. Values are expressed as arbitrary units. In all brain regions and at all time points considered, p52ShcA > p46ShcA > p66ShcA. (C) Cell lysates were obtained from undifferentiated (6 hr) and differentiated primary neuronal cultures (96 and 168 hr) generated from the E14 rat striatum primordia and grown in serum-free conditions, allowing the cells to differentiate into neurons. Ninety micrograms of protein were loaded and immunoblotted with anti-ShcA. The immunoreaction to α-tubulin was used as an internal standard. As in B, the relative densities of the immunoreactive bands are plotted after normalization over α-tubulin content and the values expressed in arbitrary units. Data shown were from one of three sets of experiments, all generating the same results.
ShcA Expression Declines During in Vitro Neuronal Differentiation.
To correlate the observed reduction in ShcA levels with the acquisition of a differentiated state, cells from the E14 rat striatum primordia were isolated and allowed to differentiate in vitro (34, 44). Previous reports showed that when exposed to serum-free conditions (SFM) the majority of the E14 striatal cells develop into neurons within 7 days (34, 44). Lysates from primary striatal cells cultured for 6, 96, and 168 hr (7 days) in SFM express progressively reduced levels of ShcA proteins (Fig. 1C). The relative densities of the immunoreactive bands are plotted after normalization over an α-tubulin signal. The correlation of the in vivo and in vitro findings of a reduced expression of ShcA proteins in coincidence with cell differentiation suggests that ShcA proteins may be of fundamental importance at early stages of neurogenesis, when there is an enhanced proliferation of the neuroblasts lining the lumen of the neural tube.
ShcA mRNAs Are Localized Within the Ventricular Zone (VZ).
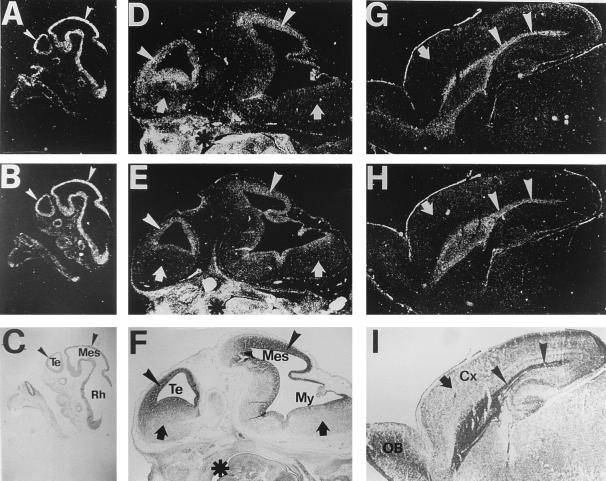
When expression of the mRNAs encoding for the three ShcA isoforms was analyzed at different developmental stages, a strong hybridization signal along the entire neural tube in E10.5 mouse embryos was revealed with two different antisense riboprobes, one recognizing all three mRNAs transcribed from the ShcA gene (ShcA riboprobe), the other specific for p66 (p66shcA riboprobe) (Fig. 2 A and B). At this stage, proliferation was exclusive to the proencephalon wall. At E12.5 (Fig. 2 D and E), actively proliferating structures like the ganglionic eminences and the VZ of the cortex (arrowheads) were still highly positive for ShcA and p66shcA. On the other hand, post mitotic areas (arrows) showed no detectable hybridization signal. Outside the brain the hybridization signal appeared to be stronger and widespread (Fig. 2, asterisks), consistent with the literature data that indicate a ubiquitous expression pattern of ShcA in extraneuronal tissues (20, 24–28). At later stages (e.g., P2) mRNA expression became restricted to the extremely small proliferative zone of the cerebral hemisphere and the striatum (arrowheads in Fig. 2 G–I). Northern analyses confirmed a reduced presence of ShcA mRNAs in the postnatal rat brain (not shown). In situ hybridization assay performed with P66 and Shc sense probes gave no specific signal (not shown).
Figure 2.
ShcA and p66ShcA localization in mouse sagittal sections. ShcA (A, D, and G) and p66ShcA (B, E, and H) expression in mouse sagittal sections. Anterior is to the left. (A–C) E10.5, (D–F) E12, (G–I) P2. Bright-field views of sections counterstained with cresyl violet are also shown (C, F, and I). Both ShcA and p66ShcA showed developmentally regulated hybridization signal exclusively in the brain. Their patterns of expression appeared to be temporally regulated and practically coincide with the proliferative epithelium (ventricular zone, VZ) all throughout development (arrowheads). p66ShcA mRNA was expressed in all territories showing the ShcA signal, although to a lesser extent as expected from the relative levels of the three isoforms. Postmitotic regions (arrows) showed no specific hybridization signal. On the contrary, outside the brain Shc and p66shcA expression patterns appear to be both ubiquitous and homogenous (asterisks in D–F). Sense probes gave no specific hybridization signals (data not shown). Cx, cortex; Mes, mesencephalon; My, myelencephalon; Rh, rhombencephalon; Te, telencephalon.
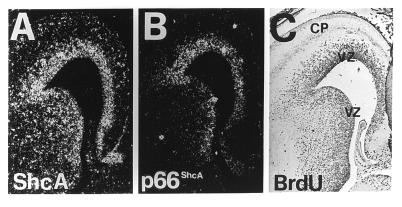
To correlate the expression of ShcA and p66shcA mRNAs with cell proliferation in the VZ, we used BrdUrd incorporation followed by immunohistochemistry (37, 38). BrdUrd is incorporated by all cells that are in the S-phase of the cell cycle at the time of the injection. Coronal sections of E14.5 brains showed ShcA and p66shcA mRNAs (Fig. 3 A and B) in the VZ, where the majority of the BrdUrd-labeled cells were located (Fig. 3C).
Figure 3.
ShcA and p66ShcA expression in developing mouse cortex. ShcA (A) and p66ShcA (B) expression in adjacent coronal sections of the developing mouse cortex at E14.5. (C) BrdUrd antibody staining after a 15-hr BrdUrd labeling. Both ShcA and p66ShcA expression patterns correlated with the position of the proliferative epithelium in the developing cortex, as revealed by BrdUrd labeling. CP, cortical plate; VZ, ventricular zone
Expression of ShcA Proteins in the Adult Nervous System Is Uniquely Localized to the Olfactory Epithelium.
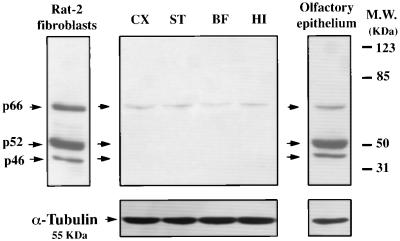
In a manner consistent with our data showing selective expression of ShcA in the proliferating neuroblasts, lysates from different regions of the adult brain presented a barely detectable amount of ShcA proteins (Fig. 4). Instead, lysates obtained from the adult olfactory epithelium, a structure where neuronal cell renewal is also known to occur in adult life (45), showed levels of ShcA proteins comparable to those observed in all of the other brain regions analyzed during early embryonic stages (Fig. 4).
Figure 4.
ShcA expression in different adult brain regions and in the olfactory epithelium. Western blot analyses showed low levels of expression in all brain regions. p66 was the only isoform present at a low, although detectable, level. On the other hand, in the olfactory epithelium all three isoforms were strongly expressed and exhibited the same relative proportional abundance as found in brain regions at early developmental stages.
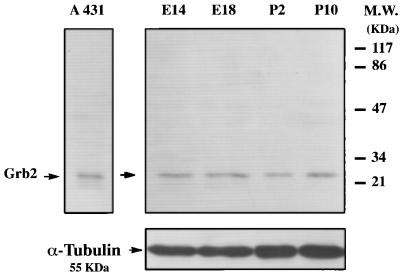
Grb2 Is Constantly Expressed Throughout Neurogenesis.
In addition to associating with activated receptor tyrosine kinases, phosphorylated ShcA proteins have been shown to form complexes with the Grb2 adaptor molecule in different cellular systems in response to specific stimuli (5, 6, 10). Grb2 adaptor protein is known to interact with activated TK receptors directly or through ShcA. The finding that ShcA was developmentally regulated prompted us to analyze whether Grb2 was expressed in the rodent forebrain at different development stages. Western blot analyses, performed on homogenates obtained from striatal tissue at different gestational days, showed constant levels of Grb2 protein all throughout neurogenesis (Fig. 5). The same results were obtained using tissue homogenates from cortex, hippocampus, and basal forebrain taken at different stages of neurogenesis (data not shown).
Figure 5.
Expression of Grb2 protein during neurogenesis. (Right) Western blot analysis showing Grb2 protein levels in lysates from striatum. Constant levels of expression were found. Samples and membranes were treated as described in Fig. 1.
In Vivo Functional Activation of Embryonic ShcA Proteins.
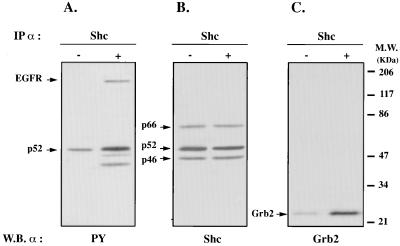
To investigate whether ShcA proteins in the VZ were subjected of being activated, we analyzed the extent of induced ShcA phosphorylation following injection of specific polypeptide growth factors into the cerebral ventricular system of rat embryos at 15 gestational days (36). Lysates from EGF-treated and untreated (control) animals were subjected to immunoprecipitation with anti-ShcA antibodies, followed by Western blot analysis with anti-phosphotyrosine antibodies. Fig. 6 shows a basal level of p52shcA phosphorylation in lysates from control animals. These data may reflect ShcA activation by endogenous stimuli (mitogens and/or differentiative factors). On the other hand, when an equal amount of lysed telencephalic material obtained from EGF-injected embryos was subjected to the same immunoprecipitation procedure, p52shcA phosphorylation was strongly induced (Fig. 6A). In Fig. 6B, the same membrane filter as in A was stripped and reacted with anti-ShcA mAb. As shown (arrows), ShcA proteins were immunoprecipitated to the same extent in control and treated groups. The finding of in vivo EGF-induced p52shcA phosphorylation is further substantiated by the presence of a 170-kDa phosphorylated band in the EGF-treated lane (Fig. 6A, arrow), which we found to react with a mAb against the EGF receptor (EGFR), which is known to coprecipitate with phosphorylated ShcA proteins (not shown) (20). Furthermore, since the activated ShcA proteins normally interact with the Grb2 adaptor protein and we found Grb2 to be present constantly throughout development, we evaluated the presence of Grb2 in lysates from control and EGF-stimulated groups that had been subjected to immunoprecipitation with anti-ShcA antibodies. As shown in Fig. 6C, the 23-kDa Grb2 protein coprecipitates with anti-ShcA antibodies in lysates from treated embryos.
Figure 6.
p52ShcA was phosphorylated and interacted with Grb2 in the embryonic telencephalic vesicles following intraventricular injection of EGF. (A) Phosphotyrosine immunoblot of ShcA immunoprecipitates after in vivo EGF treatment; 10 min after injection, the embryos were removed and the telencephalic vesicles isolated and prepared for the immunoprecipitation. Phosphorylated p52ShcA is indicated. Control embryos were injected with vehicle. A 170-kDa phosphorylated band (arrow) corresponding to the coprecipitated EGF receptor is also visible in the treated group. (B) Anti-ShcA immunoblot of the filter shown in A. The membrane was stripped and reacted with ShcA mAb. As shown in the figure, ShcA proteins were immunoprecipitated to the same extent in control and EGF-treated groups. (C) Anti-Grb2 immunoblot of the same immunoprecipitates as in A. Arrow indicates the 23-kDa Grb2 protein, which more abundantly coprecipitates with ShcA in the treated group.
DISCUSSION
Several previous studies have suggested a ubiquitous expression of ShcA adaptor proteins in different nonneuronal cell types, independent of the stage of maturation of the tissue considered, where they act as key regulators of cell responsiveness to extracellular signals. In this paper, however, we found that in the mammalian brain ShcA expression is strictly regulated both temporally and spatially during embryonic development.
Surprisingly, we found that the pattern of expression of this adaptor protein correlates with the proliferative and early differentiative events that occur in the brain, supporting the hypothesis that ShcA proteins may have an important role in neural development. This finding was further confirmed by in vitro studies demonstrating a reduced level of expression of ShcA isoforms during neuronal differentiation.
To establish a direct correlation between the presence of ShcA and brain neurogenesis, we examined the localization of ShcA mRNAs at different developmental stages. It is known that during brain development active proliferation of neuroblasts occurs in an anatomically defined and highly organized layer lining the lumen of the neural tube, called the VZ (46). Subsequently, at given intervals and depending on the species considered, these cells differentiate into neurons following spatially defined gradients of neurogenesis, which are specific for each brain region. In the mouse cortex, for example, this phenomenon starts at E11 (47, 48) and the first postmitotic cells migrate away from the lumen forming the so-called cortical plate, which will later give rise to the mature six-layered cerebral cortex (43). A similar temporally regulated and spatially defined departure of postmitotic cells from the VZ is also known to occur in other brain regions (49, 50).
Our analyses showed that, in the brain, expression of ShcA adaptor proteins is exclusively restricted to the VZ, where they are susceptible to activation by stimulation with exogenous factors. With the occurrence of cell differentiation, a change in the levels of ShcA proteins occurs. In fact, in the postnatal brain, ShcA mRNA expression remains confined to the subependymal layer lining the ventricular system. In the adult brain, the three ShcA isoforms are only faintly detectable. This finding is in agreement with recent reports that found low levels of expression of ShcA in total mouse brain extracts (32, 33). Interestingly, we found that ShcA proteins remain strongly expressed in the adult olfactory epithelium in which neuronal cell renewal occurs throughout life. All these phenomena were peculiar to ShcA. In fact, when the presence of another adaptor protein, Grb2, was assessed, no particular changes in expression levels were observed during neurogenesis. These data, therefore, argue in favor of an important role for ShcA at the transition from proliferation to differentiation in the brain.
These findings indicate that (i) ShcA adaptor proteins are subjected to strict developmental control, their expression being associated with the proliferative events of the developing mammalian forebrain and adult olfactory epithelium and (ii) the mRNAs that give rise to the three ShcA isoforms are expressed in a spatially restricted manner and confined to an anatomically defined region of the developing brain, i.e., the germinal epithelium, a transitory structure known to include the population of proliferating neuroblasts.
These data support the hypothesis that important variations in ShcA levels during neurogenesis may change the responses of a particular cell type to external stimuli. Loss of ShcA during neurogenesis may guarantee a cell from undesired proliferation, and/or may represent the major force that drives neuroblasts from proliferation into differentiation.
In mature brain cells, the Ras signaling pathway may be activated by proteins different from ShcA. In this respect, the two recently isolated Shc proteins, ShcB and ShcC, have been found to be expressed in total extracts from the adult brain (32, 33) and our preliminary data indicate that their expression levels increase in association with the decrease in ShcA protein levels during development (E.C., unpublished work).
We found that ShcA proteins expressed in neuroblasts can be activated, since after EGF injection into the cerebral ventricular system of rat embryos at 15 gestational days, an increased phosphorylation of p52shcA isoform was detected by immunoprecipitation and Western blot analyses. Other in vitro investigations have reported a similar prevalence of phosphorylated p52shcA in stimulated cells (20, 24–28). We focused on EGF since (i) this molecule shares, with other growth factors, the ability to elicit proliferation of immature mammalian central nervous system progenitor cells (1–3, 51–55) and (ii) in nonneuronal cellular systems activation of its corresponding surface receptor has been shown to induce ShcA phosphorylation (11, 20, 24–29). Furthermore, in addition to associating with activated receptor tyrosine kinases, phosphorylated ShcA proteins have been shown to form complexes with the Grb2 adaptor molecule in different cellular systems in response to specific stimuli (4–6, 13, 14). To further consolidate our results indicating ShcA recruitment in vivo in the population of proliferating neuroblasts lining the neural tube, we showed that Grb2 coprecipitates with the immunoprecipitated ShcA proteins in EGF-stimulated embryonic brains.
These results, showing in vivo EGF-induced p52shcA phosphorylation and its association to Grb2, combined with the data on the restricted expression of ShcA proteins in the VZ, indicate functional activity of ShcA proteins in the embryonic neuroepithelium.
Although we have focused mainly on ShcA contribution from neuroblasts, proliferating glial elements populating the brain at advanced stages of development and reactive astrocytes found in the adult injured brain may also express ShcA.
We believe that the findings reported here define new modalities by which the switch between proliferation and differentiation of immature neuroblasts may be controlled, suggesting that differences in the levels of ShcA adaptor proteins at different stages of the development of a brain cell may change the pattern of protein–protein interactions induced by activation of a particular membrane receptor, thereby setting that cell to respond differently to the same extracellular factor. From this perspective, the fact that levels of ShcA proteins vary according to the cell state may be of particular relevance since these proteins act as initiators of a signaling cascade that links growth factor polypeptides to nuclear events.
The possibility that cells that have to escape from the VZ might modify their adaptor proteins such that the extracellular interactions will promote cell migration/differentiation rather than preventing it, and/or activate new genes in the cascade of differentiation, remains an intriguing possibility.
Acknowledgments
We would like to thank Pierre Magistretti, Lorenzo Magrassi, Rodolfo Paoletti, Pier Giuseppe Pelicci, Larry Wrabetz, and Vincenzo Zappavigna for helpful discussion and comments on the manuscript, and Lorenzo Magrassi for the suggestion to test the adult olfactory epithelium. This research was supported by Associazione Italiana Ricerca sul Cancro (442/96), Telethon (A71), North Atlantic Treaty Organization–Collaborative Research Group (969294) and a grant from the Huntington’s Disease Society of America to E.C.
ABBREVIATIONS
- TK
thymidine kinase
- EGF
epidermal growth factor
- E
embryonic day
- VZ
ventricular zone
References
- 1.Gage F H, Ray J, Fisher L J. Annu Rev Neurosci. 1995;18:159–192. doi: 10.1146/annurev.ne.18.030195.001111. [DOI] [PubMed] [Google Scholar]
- 2.Temple S, Qian X. Curr Opin Neurobiol. 1996;6:11–17. doi: 10.1016/s0959-4388(96)80003-1. [DOI] [PubMed] [Google Scholar]
- 3.Henderson C E. Curr Opin Neurobiol. 1996;6:64–70. doi: 10.1016/s0959-4388(96)80010-9. [DOI] [PubMed] [Google Scholar]
- 4.Schlessinger J, Ullrich A. Neuron. 1992;9:383–391. doi: 10.1016/0896-6273(92)90177-f. [DOI] [PubMed] [Google Scholar]
- 5.Fantl W J, Johnson D E, Williams L T. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- 6.van der Geer P, Hunter T, Lindberg R A. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 7.Buday L, Downward J. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 8.Egan S E, Giddings B W, Brooks M W, Buday L, Sizeland A M, Weinberg R A. Nature (London) 1993;363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- 9.Li N, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Nature (London) 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 10.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 11.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ullrich A, Skolnik E Y, Bar-Sagi D, Schlessinger J. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 12.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Botwell D. Nature (London) 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 13.Bonfini L, Migliaccio E, Pelicci G, Lanfrancone L, Pelicci P G. Trends Biochem Sci. 1996;21:257–261. [PubMed] [Google Scholar]
- 14.Waters S B, Chen D, Kao A W, Okada S, Holt K H, Pessin J E. J Biol Chem. 1996;271:18224–18230. doi: 10.1074/jbc.271.30.18224. [DOI] [PubMed] [Google Scholar]
- 15.McGlade J, Cheng A, Pelicci G, Pelicci P G, Pawson T. Proc Natl Acad Sci USA. 1992;89:8869–8873. doi: 10.1073/pnas.89.19.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns L A, Karnitz L M, Sutor S L, Abraham R T. J Biol Chem. 1993;268:17659–17661. [PubMed] [Google Scholar]
- 17.Damen J E, Liu L, Kutler R L, Krystal G. Blood. 1993;82:2269–2303. [PubMed] [Google Scholar]
- 18.Dilworth S M, Brewster C E, Jones M D, Lanfrancone L, Pelicci G, Pelicci P G. Nature (London) 1994;367:87–90. doi: 10.1038/367087a0. [DOI] [PubMed] [Google Scholar]
- 19.Lanfrancone L, Pelicci G, Brizzi M F, Casciari C, Giuli S, Pergoraro L, Pawson T, Pelicci P G. Oncogene. 1995;10:907–917. [PubMed] [Google Scholar]
- 20.Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Grignani F, Pawson T, Pelicci P G. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- 21.Blaikie P, Immanuel D, Wu J, Li N, YaJ̇nik V, Margolis B. J Biol Chem. 1994;269:32031–32034. [PubMed] [Google Scholar]
- 22.Kavanaugh W M, Williams L T. Science. 1994;266:1862–1865. doi: 10.1126/science.7527937. [DOI] [PubMed] [Google Scholar]
- 23.van der Geer P, Pawson T. Trends Biochem Sci. 1995;20:277–280. doi: 10.1016/s0968-0004(00)89043-x. [DOI] [PubMed] [Google Scholar]
- 24.Cantrell D. Annu Rev Immunol. 1996;14:259–274. doi: 10.1146/annurev.immunol.14.1.259. [DOI] [PubMed] [Google Scholar]
- 25.Kharbanda S, Saleem A, Yuan Z, Emoto Y, Prasad K V, Kufe D. Proc Natl Acad Sci USA. 1995;92:6132–6136. doi: 10.1073/pnas.92.13.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osman N, Lucas S C, Turner H, Cantrell D. J Biol Chem. 1995;270:13981–13986. doi: 10.1074/jbc.270.23.13981. [DOI] [PubMed] [Google Scholar]
- 27.Raff-Jamison S, McGlade J, Pawson T, Chen K, Cohen S. J Biol Chem. 1993;268:7610–7612. [PubMed] [Google Scholar]
- 28.Pronk G J, McGlade J, Pelicci G, Pawson T, Bos J L. J Biol Chem. 1993;268:5748–5753. [PubMed] [Google Scholar]
- 29.Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thomas S, Brugge J, Pelicci P G, Schlessinger J, Pawson T. Nature (London) 1992;360:689–692. doi: 10.1038/360689a0. [DOI] [PubMed] [Google Scholar]
- 30.Migliaccio E, Mele S, Salcini A E, Pelicci G, Lai K-M V, Superti-Furga G, Pawson T, Di Fiore P P, Lanfrancone L, Pelicci P P. EMBO J. 1997;16:706–716. doi: 10.1093/emboj/16.4.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai K M, Olivier J P, Gish G D, Henkemeyer M, McGlade J, Pawson T. Mol Cell Biol. 1995;15:4810–4818. doi: 10.1128/mcb.15.9.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Bryan J P, Songyang Z, Cantley L, Der C J, Pawson T. Proc Natl Acad Sci USA. 1996;93:2729–2734. doi: 10.1073/pnas.93.7.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelicci G, Dente L, De Giuseppe A, Verducci-Galletti B, Giuli S, Mele S, Vetriani C, Giorgio M, Pandolfi P, Cesareni G, Pelicci P G. Oncogene. 1996;13:633–641. [PubMed] [Google Scholar]
- 34.Cattaneo E, McKay R. Nature (London) 1990;347:762–765. doi: 10.1038/347762a0. [DOI] [PubMed] [Google Scholar]
- 35.Cattaneo E, De Fraja C, Conti L, Reinach B, Bolis L, Govoni S, Liboi E. J Biol Chem. 1996;271:23374–23379. doi: 10.1074/jbc.271.38.23374. [DOI] [PubMed] [Google Scholar]
- 36.Cattaneo E, Magrassi L, Butti G, Santi L, Giavazzi A, Pezzotta S. Dev Brain Res. 1994;83:197–208. doi: 10.1016/0165-3806(94)00137-5. [DOI] [PubMed] [Google Scholar]
- 37.Gratzner H G. Science. 1982;218:474–475. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi T, Nowakowski R S, Caviness V S., Jr J Neurocytol. 1992;21:185–197. doi: 10.1007/BF01194977. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi T, Nowakowski R S, Caviness V S., Jr J Neurosci. 1993;13:820–833. doi: 10.1523/JNEUROSCI.13-02-00820.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkinson D G. In Situ Hybridization: A Practical Approach. Oxford, U.K.: IRL; 1992. [Google Scholar]
- 41.Gulisano M, Broccoli V, Pardini C, Boncinelli E. Eur J Neurosci. 1996;8:1037–1050. doi: 10.1111/j.1460-9568.1996.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 42.Simmons D M, Arriza J L, Swanson L W. J Histotechnol. 1989;12:169–181. [Google Scholar]
- 43.Bayer S A, Altman J. Neocortical Development. New York: Raven; 1991. [Google Scholar]
- 44.Weiss S, Pin J P, Sebben M, Kemp D E, Sladeczek F, Gabrion J, Bockaert J. Proc Natl Acad Sci USA. 1986;83:2238–2242. doi: 10.1073/pnas.83.7.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graziadei P P C, Monti Graziadei G A. In: Handbook of Sensory Physiology. Jacobson M, editor. Vol. 9. Berlin: Springer; 1978. pp. 55–83. [Google Scholar]
- 46.Boulder Committee. Anat Rec. 1970;16:257–262. doi: 10.1002/ar.1091660214. [DOI] [PubMed] [Google Scholar]
- 47.Angevine J B, Jr, Sidman R L. Nature (London) 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 48.Caviness V S., Jr Dev Brain Res. 1982;4:293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- 49.Hatten M E, Heintz N. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 50.Smart I, Sturrock R. In: The Neostriatum. Divak I, Oberg R, editors. New York: Pergamon; 1979. pp. 127–146. [Google Scholar]
- 51.Anchan R M, Reh T, Angello J, Balliet A, Walker M. Neuron. 1991;6:923–936. doi: 10.1016/0896-6273(91)90233-p. [DOI] [PubMed] [Google Scholar]
- 52.Reynolds B A, Weiss S. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 53.Reynolds B, Tetzlaff W, Weiss S. J Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vescovi A L, Reynolds B A, Fraser D D, Weiss S. Neuron. 1993;11:951–966. doi: 10.1016/0896-6273(93)90124-a. [DOI] [PubMed] [Google Scholar]
- 55.Svendsen C N, Fawcett J W, Bentlage C, Dunnett S B. Exp Brain Res. 1995;102:407–414. doi: 10.1007/BF00230645. [DOI] [PubMed] [Google Scholar]