Abstract
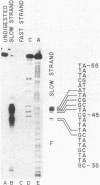
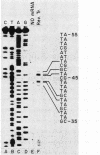
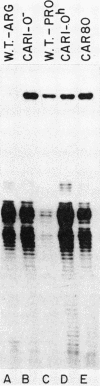
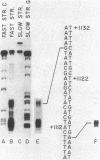
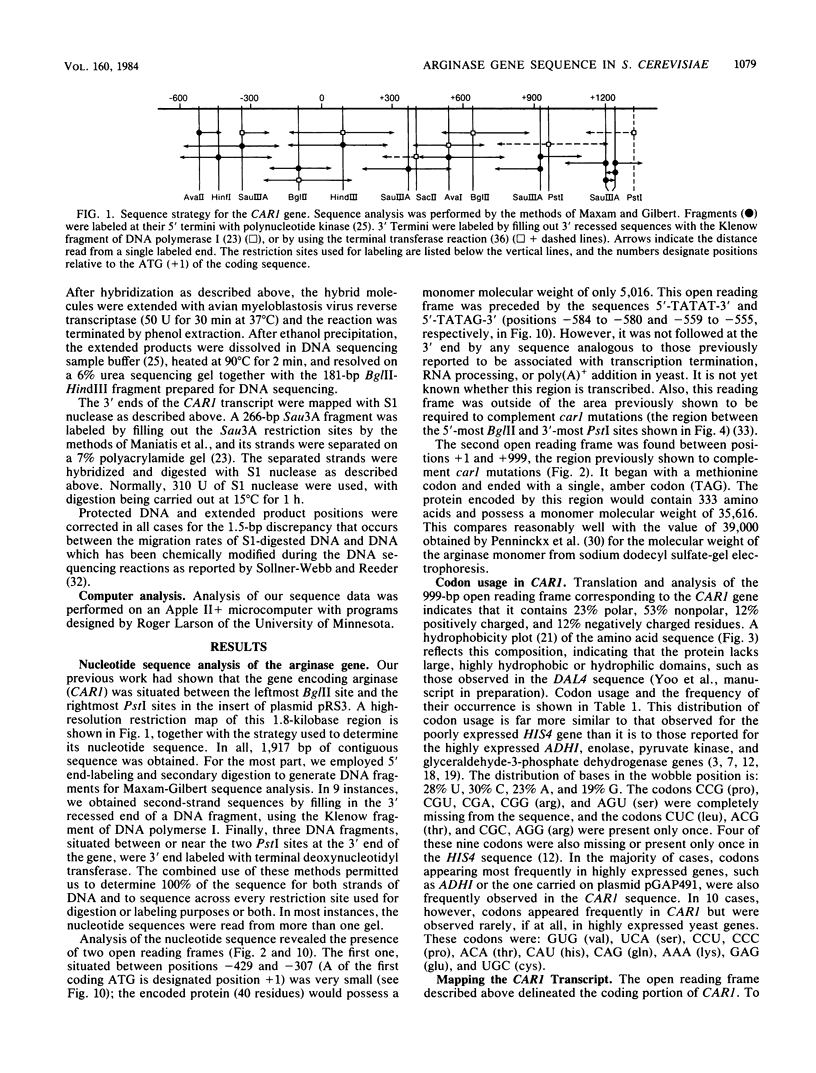
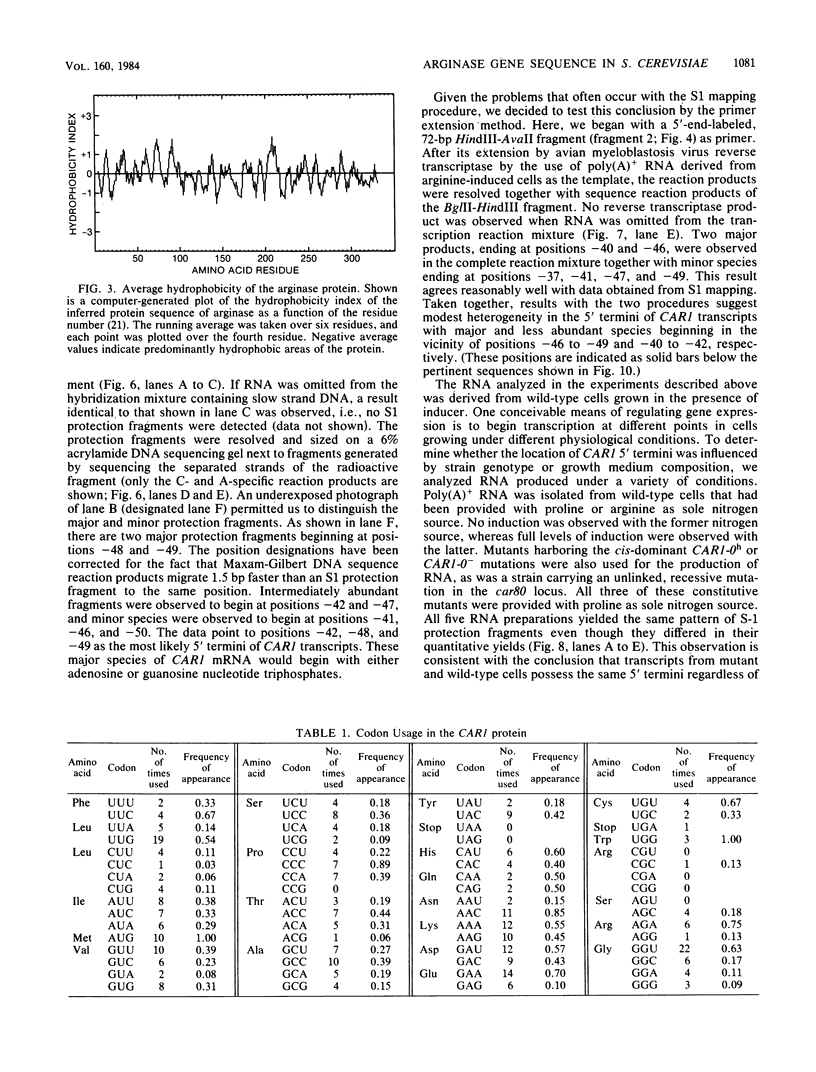
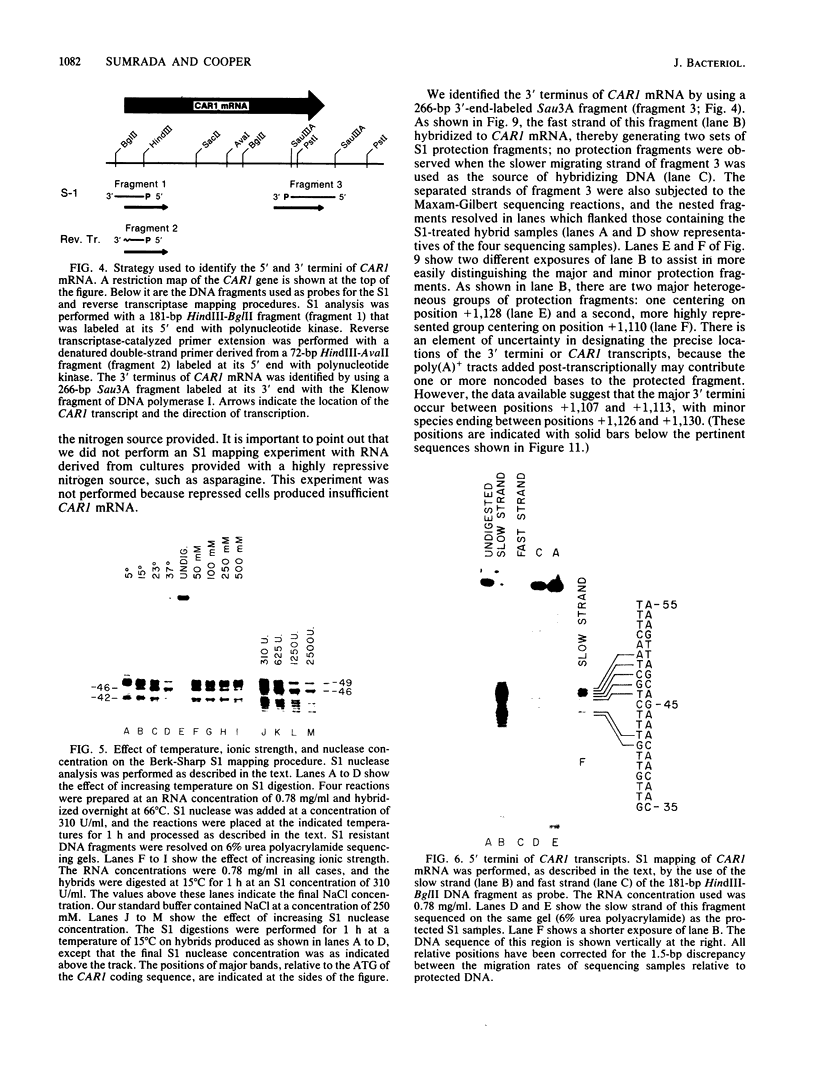
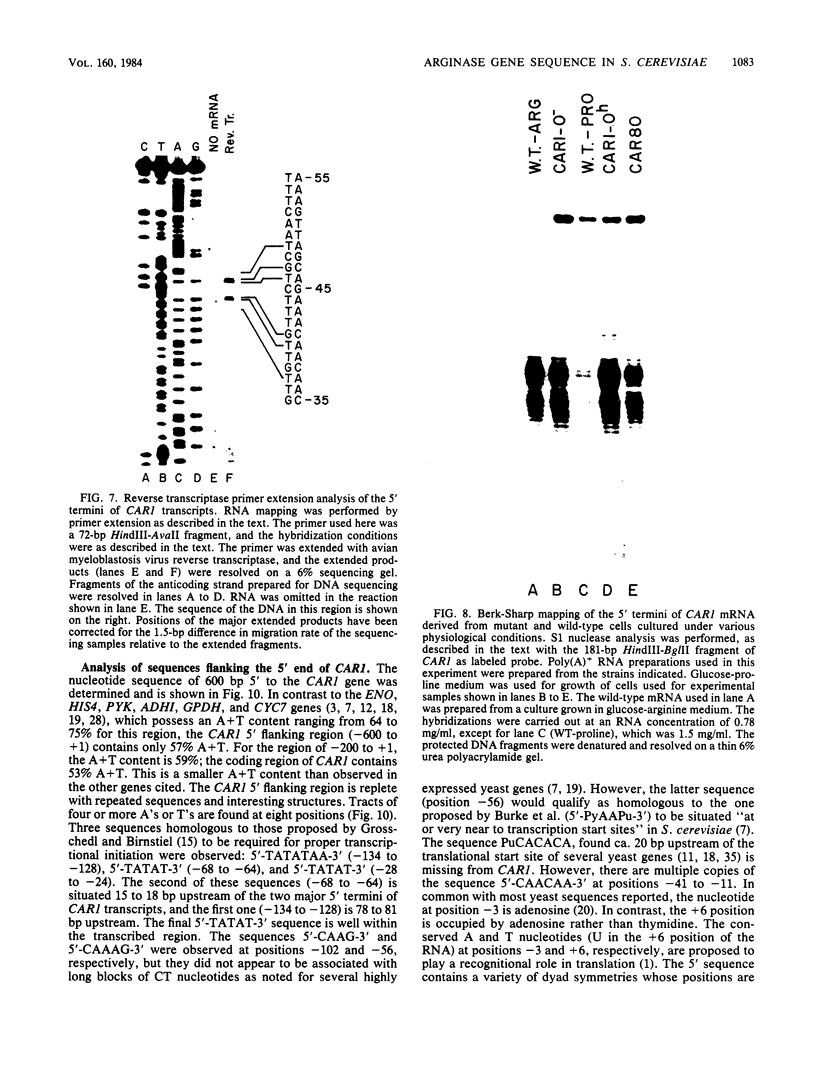
We have determined the nucleotide sequence of the yeast CAR1 gene along with its 5' and 3' flanking sequences. The structural portion of this gene encodes a protein of 333 amino acid residues with a calculated Mr value of 35,616. The transcripts produced from this gene are modestly heterogeneous at their 5' and 3' termini. Most 5' termini map to a position 42 to 49 base pairs upstream from the ATG at the beginning of the open reading frame. The 3' termini map to a position 108 to 127 base pairs downstream of the amber codon which terminates the open reading frame. A variety of potentially significant sequences were identified in the regions flanking the structural portion of the gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bechet J., Greenson M., Wiame J. M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970 Jan;12(1):31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossinger J., Cooper T. G. Molecular events associated with induction of arginase in Saccharomyces cerevisiae. J Bacteriol. 1977 Jul;131(1):163–173. doi: 10.1128/jb.131.1.163-173.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Crabeel M., Huygen R., Cunin R., Glansdorff N. The promoter region of the arg3 gene in Saccharomyces cerevisiae: nucleotide sequence and regulation in an arg3-lacZ gene fusion. EMBO J. 1983;2(2):205–212. doi: 10.1002/j.1460-2075.1983.tb01406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Dubois E., Hiernaux D., Grennon M., Wiame J. M. Specific induction of catabolism and its relation to repression of biosynthesis in arginine metabolism of Saccharomyces cerevisiae. J Mol Biol. 1978 Jul 15;122(4):383–406. doi: 10.1016/0022-2836(78)90417-5. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Hoet P. P., Wiame J. M. On the nature of argR mutations is Saccharomyces cerevisiae. Eur J Biochem. 1974 Mar 15;43(1):87–92. doi: 10.1111/j.1432-1033.1974.tb03388.x. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. The primary structure of a glyceraldehyde-3-phosphate dehydrogenase gene from Saccharomyces cerevisiae. J Biol Chem. 1979 Oct 10;254(19):9839–9845. [PubMed] [Google Scholar]
- Holland M. J., Holland J. P., Thill G. P., Jackson K. A. The primary structures of two yeast enolase genes. Homology between the 5' noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981 Feb 10;256(3):1385–1395. [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee D. C., Roeder R. G. Transcription of adenovirus type 2 genes in a cell-free system: apparent heterogeneity of initiation at some promoters. Mol Cell Biol. 1981 Jul;1(7):635–651. doi: 10.1128/mcb.1.7.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D. J., Chambon P. The SV40 early region TATA box is required for accurate in vitro initiation of transcription. Nature. 1981 Mar 26;290(5804):310–315. doi: 10.1038/290310a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. Participation of transcriptional and post-transcriptional regulatory mechanisms in the control of arginine metabolism in yeast. Mol Gen Genet. 1983;189(1):148–156. doi: 10.1007/BF00326068. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K., Hall B. D., Astell C., Smith M. Physical analysis of mating-type loci in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):961–981. doi: 10.1101/sqb.1981.045.01.113. [DOI] [PubMed] [Google Scholar]
- Penninckx M., Simon J. P., Wiame J. M. Interaction between arginase and L-ornithine carbamoyltransferase in Saccharomyces cerevisiae. Purification of S. cerevisiae enzymes and evidence that these enzymes as well as rat-liver arginase are trimers. Eur J Biochem. 1974 Nov 15;49(2):429–442. doi: 10.1111/j.1432-1033.1974.tb03848.x. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Tu C. P., Cohen S. N. 3'-end labeling of DNA with [alpha-32P]cordycepin-5'-triphosphate. Gene. 1980 Jul;10(2):177–183. doi: 10.1016/0378-1119(80)90135-3. [DOI] [PubMed] [Google Scholar]
- Whitney P. A., Magasanik B. The induction of arginase in Saccharomyces cerevisiae. J Biol Chem. 1973 Sep 10;248(17):6197–6202. [PubMed] [Google Scholar]
- Wickerham L. J. A Critical Evaluation of the Nitrogen Assimilation Tests Commonly Used in the Classification of Yeasts. J Bacteriol. 1946 Sep;52(3):293–301. [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]