Abstract
The molecular evolution of the opioid receptor family has been studied by isolating cDNAs that encode six distinct opioid receptor-like proteins from a lower vertebrate, the teleost fish Catostomus commersoni. One of these, which has been obtained in full-length form, encodes a 383-amino acid protein that exhibits greatest sequence similarity to mammalian μ-opioid receptors; the corresponding gene is expressed predominantly in brain and pituitary. Transfection of the teleost cDNA into HEK 293 cells resulted in the appearance of a receptor having high affinity for the μ-selective agonist [d-Ala2, MePhe4-Gly-ol5]enkephalin (DAMGO) (Kd = 0.63 ± 0.15 nM) and for the nonselective antagonist naloxone (Kd = 3.1 ± 1.3 nM). The receptor had negligible affinity for U50488 and [d-Pen2, d-Pen5]enkephalin (DPDPE), which are κ- and δ-opioid receptor selective agonists, respectively. Stimulation of transfected cells with 1 μM DAMGO lowered forskolin-induced cAMP levels, an effect that could be reversed by naloxone. Experiments in Xenopus oocytes have demonstrated that the fish opioid receptor can, in an agonist-dependent fashion, activate a coexpressed mouse G-protein-gated inward-rectifying potassium channel (GIRK1). The identification of six distinct fish opioid receptor-like proteins suggests that additional mammalian opioid receptors remain to be identified at the molecular level. Furthermore, our data indicate that the μ-opioid receptor arose very early in evolution, perhaps before the appearance of vertebrates, and that the pharmacological and functional properties of this receptor have been conserved over a period of ≈400 million years implying that it fulfills an important physiological role.
Keywords: cDNA cloning, functional expression, peptide receptor, teleost fish, Xenopus oocyte
Opioid receptors mediate the effects of morphine and its derivatives and are the targets of endogenous opioid peptides. They have been pharmacologically classified into three major categories (μ, δ, and κ), each containing at least two subtypes (1–3). cDNAs that encode mammalian δ- (4–6), κ- (6–9), and μ-receptors (10–13) have been described. In addition, cDNA cloning (14, 15) has revealed the existence of an opioid receptor-like protein (ORL1), which has been shown to bind an endogenous peptide that has been called nociceptin (16) and orphanin FQ (17). All of these receptors are members of the G-protein-coupled receptor superfamily and, thus, are each predicted to contain seven transmembrane domains. Activation of opioid receptors results, through a pertussis-toxin sensitive mechanism, in the inhibition of adenylate cyclase and the modulation of calcium and potassium conductances (3, 18).
The μ-opioid receptor mediates the analgesic and addictive effects of morphine, and mice deficient for this receptor do not respond to this drug in tests for antinociception and physical dependence (19); somewhat surprisingly, these mice exhibit no morphological or behavioral abnormalities. Although the physiological function(s) of the μ-opioid receptor (as well as of other opioid receptors) remains unclear, it has been suggested that the opioid system is inactive under normal resting conditions and is only called into action in response to particular environmental circumstances (20). In an attempt to shed light on the biological roles played by opioid receptors, and to investigate their phylogeny, we have isolated cDNAs from a teleost fish, namely the white sucker Catostomus commersoni. We show here that the sequence, pharmacology, second messenger coupling, and agonist-induced potassium channel (GIRK1; G-protein-gated inward-rectifying potassium channel) response of one of these (a μ-receptor) have been highly conserved during the course of vertebrate evolution.
MATERIALS AND METHODS
Cloning of C. commersoni Opioid Receptor cDNAs.
Random hexamer-primed cDNA, synthesized from total RNA isolated from the brain of C. commersoni, was amplified using the degenerate oligonucleotide primers: 5′-AAGAGATCTACXAA(C/T)AT(A/C/T)TA(C/T)AT(A/C/T)TT(C/T)AA-3′, where X = A, C, G, and T, and 5′-GGGGAGCTCACXGCCATXACCAT(A/G/T)ATXGG-3′. These recognize the nucleotide sequences that encode amino acids 81–90 (KTATNIYIFN; single-letter code) and 182–191 (PIMVMAVTNP), respectively, of the mouse δ-opioid receptor (5). Products were cloned into M13 vectors, taking advantage of restriction endonuclease recognition sites (underlined) incorporated into the 5′ ends of the PCR primers, and sequenced. To obtain the 5′ and 3′ sequences that were missing from one of the partial clones, we applied the rapid amplification of cDNA ends technique (21) to C. commersoni whole brain first-strand cDNA, essentially as described (22, 23). These sequences were used to design two primers: 5′-TGTGAATTCTGGGCTGGCAGCGCGACTCCA-3′ and 5′-TTCGAATTCGGACAGTCCTGACATCGAAGC-3′, which correspond to nucleotide sequences that flank the coding region. The primers were employed in the PCR to generate a full-length cDNA, which was cloned into pBluescript KS+, yielding plasmid psuck10, and sequenced.
Transfection and Ligand-Binding Assays.
An ≈1.2-kb EcoRI–XhoI fragment of psuck10, which contains the entire open reading frame, was subcloned into the expression vector pcDNA3 (Invitrogen) downstream of the cytomegalovirus (CMV) promoter. Human embryonic kidney 293 (HEK 293) cells (American Type Culture Collection) were transfected using the calcium phosphate method (24), and stable transfectants were selected in the presence of 450 μg/ml geneticin (G-418 sulfate; GIBCO/BRL) without clonal selection (25). Membranes were prepared and binding (10 μg protein/reaction) was performed at room temperature, for 1 h, in 20 mM Hepes (pH 7.4), 3 mM MgCl2, 0.05% (wt/vol) BSA, 2 mM EGTA, 0.4 μg/ml bacitracin, 0.5 μg/ml phenylmethylsulfonyl fluoride, 0.02 μg/ml leupeptin, 4 μg/ml pepstatin, and 16 units/ml trasylol, using either [3H]DAMGO ([d-Ala2, MePhe4-Gly-ol5]enkephalin; Amersham, 58 Ci/mmol; 1 Ci = 37 GBq) or [N-allyl-2,3-3H]naloxone (Amersham, 41 Ci/mmol). Reactions were terminated by vacuum filtration through GF/C filters (Whatman) that had been pretreated with 0.5% (vol/vol) polyethylenimine. These were then washed twice with ice-cold buffer comprising 15 mM Tris⋅HCl (pH 7.4), 2 mM MgCl2, and 0.05% (wt/vol) BSA. Filter-bound radioactivity was then determined by liquid scintillation counting. Nonspecific binding was determined in the presence of 10 μM naloxone.
cAMP Assay.
Stably transfected HEK 293 cells were cultured in 24-well microtiter plates that had been pretreated with poly-d-lysine. Confluent cell monolayers were incubated with 500 μM 3-isobutyl-1-methylxanthine in DMEM (GIBCO/BRL), without serum, at 37°C for 60 min. The medium was then replaced with DMEM containing 16 nM forskolin and various opioid receptor ligands. After incubation at 37°C for 30 min, the medium was removed and the cells were kept in 70% (vol/vol) ethanol at −20°C overnight. The cell extracts were then collected and lyophilized, and then the cAMP content was determined using a radioimmunoassay kit (DuPont).
Functional Expression in Xenopus Oocytes.
RNAs were synthesized in vitro from plasmid psuck10, from a pcDNA3 plasmid containing a full-length mouse GIRK1 cDNA, and from a pRc/CMV plasmid (Invitrogen) containing a full-length rat μ-opioid receptor (MOR1a) cDNA. Opioid receptor and GIRK1 RNAs were mixed in a 1:1 ratio and injected, at a concentration of 80 ng/μl, into Xenopus laevis oocytes. Whole-cell voltage-clamp recordings were made 2–4 days after injection. Oocytes were superfused with ND-96 medium (96 mM NaCl/2 mM KCl/1.8 mM CaCl2/1 mM MgCl2/5 mM Hepes, pH 7.4) and clamped at −80 mV. To measure the effect of opioid receptor agonists, the medium was changed to a high potassium medium (hK; ND-96 but with 96 mM KCl/2 mM NaCl). After the initial potassium-activated inward current reached a plateau, agonists were applied in hK. Following washout of the agonists with hK, 300 μM BaCl2 (in hK) was applied to determine the contribution of GIRK1-mediated currents to the total inward current observed.
Northern Blot Anaysis and Reverse Transcription–PCR (RT-PCR).
Total RNAs were prepared using the RNA isolation solvent RNA-Clean (AGS, Heidelberg), and dissolved in water. Approximately 50 μg of each RNA was electrophoresed through a 0.7% (wt/vol) agarose/formaldehyde gel and transferred to a Hybond-N nylon membrane (Amersham). The blot was hybridized, in 50% (vol/vol) formamide, 4× SSC (1× SSC = 0.15 M NaCl/0.015 M sodium citrate, pH 7), 0.5% (wt/vol) SDS, 5× Denhardt’s solution, 1 mM sodium pyrophosphate, 25 mM sodium phosphate (pH 7), 100 μg/ml polyadenylic acid, 120 μg/ml heparin, and 100 μg/ml yeast tRNA, at 50°C for 18 h. An ≈820-bp EcoRI–SphI fragment from psuck10 was used as a probe; this was labeled with [α-32P]dCTP, to high specific activity, using a Prime-It II random nonamer labeling kit (Stratagene). The blot was washed twice in 0.1× SSC, 0.1% (wt/vol) SDS, at 65°C for 15 min, and then exposed to BioMax x-ray film (Kodak). The blot was subsequently stripped and rehybridized, under the same conditions, with an ≈1,070-bp BglII–EcoRI fragment of a C. commersoni β-actin cDNA (F.R.G., unpublished data), to confirm that similar amounts of RNA had been loaded in each lane.
For RT-PCR, total RNA (15 μg per sample) was treated with RNase-free DNase (Promega) and first-strand cDNA was synthesized, in a final volume of 50 μl, using random nonamers (Stratagene). Control (“sham”) reactions, from which reverse transcriptase was omitted, were set up in parallel. A total of 1 μl of each reaction was then amplified using oligonucleotides 5′-CCTGGTACCGGGAGACCCTCCTGAAGATCT-3′ and 5′-TGTGAATTCTGGGCTGGCAGCGCGACTCCA-3′ (which recognize the fish μ-opioid receptor cDNA), and 5′-CCTGAGCTCAAATACTCCGTCTGGA-3′ and 5′-TCTCTGCAGAGTTTGAGTCGGCGTG-3′ (which recognize the fish β-actin cDNA). Amplification was for either 35 (μ-receptor) or 30 (β-actin) cycles of 94°C for 1 min (denaturation), 65°C for 1 min (annealing), and 72°C for 1 min (extension). Note that no product was generated in any of the sham control amplifications.
RESULTS
Sequence of a Teleost Fish μ-Opioid Receptor.
Using degenerate oligonucleotide primers, the sequences of which were based on the mouse δ-opioid receptor sequence (5), we have isolated six different C. commersoni cDNA fragments, each of which encodes an opioid receptor-like sequence. One of these has been obtained in full-length form using the 5′ and 3′ rapid amplification of cDNA ends method (21). Sequence analysis of this cDNA revealed an open reading frame for a 43.2-kDa protein that comprises 383 amino acids (Fig. 1). The protein exhibits strong sequence similarity to the mouse, rat, and human μ-opioid receptors (75%, 75%, and 74% identity, respectively); it displays much weaker identity to the mammalian δ-, κ-, and ORL1 receptors (58%, 60%, and 52%, respectively, to the rat sequences). The teleost μ-like receptor has seven hydrophobic, putative membrane-spanning domains (TM1 to TM7), five consensus sites for the addition of N-linked sugars (Asn-6, Asn-21, Asn-28, Asn-31, and Asn-39), which are located N-terminal of TM1, and two potential palmitoylation sites (Cys-342 and Cys-347) after TM7 (Fig. 1). In addition, the sequence contains sites for phosphorylation by protein kinase A (Thr-275, between TM5 and TM6) and protein kinase C (Thr-169, between TM3 and TM4; Ser-257, between TM5 and TM6; and Ser-359, after TM7).
Figure 1.
Primary structure of the C. commersoni μ-opioid receptor. The fish μ-opioid receptor sequence has been aligned with those of the rat μ- (11), κ- (9), and δ- (11) opioid receptors, and the rat ORL1 receptor (14), using the computer program pileup (Wisconsin Sequence Analysis Package, Version 8, Genetics Computer Group, Madison, WI). The seven putative membrane-spanning domains which are common to G-protein-coupled receptors are enclosed in boxes, and asterisks mark positions at which the C. commersoni and rat μ-receptor sequences are identical. In the fish sequence, consensus N-linked glycosylation sites are boxed, and potential targets for phosphorylation by either protein kinase A or C (see text) are high-lighted by white lettering on a black background.
Pharmacology of the Teleost Fish μ-Opioid Receptor.
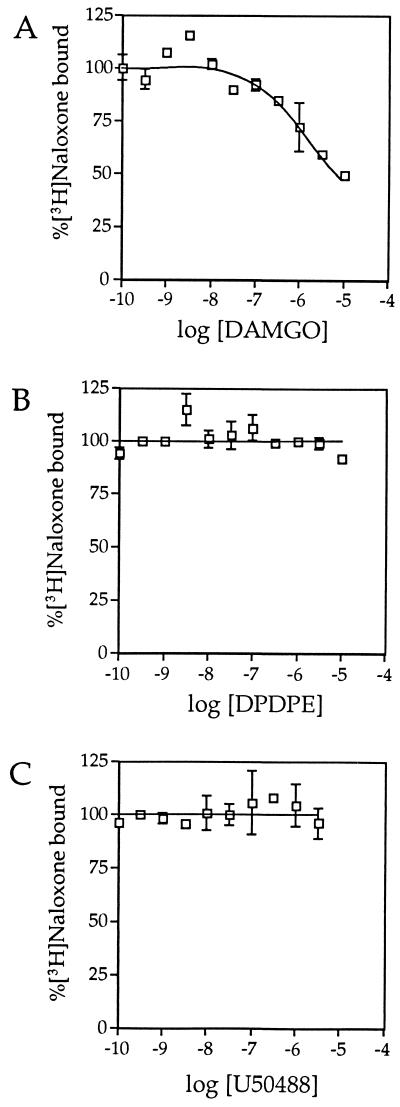
To pharmacologically characterize the C. commersoni μ-like receptor, we transfected the cloned cDNA into HEK 293 cells and performed radioligand-binding experiments on cell membranes. Using the nonselective opioid receptor antagonist [3H]naloxone, binding was observed to be saturable (Fig. 2A) and due to a single class of high-affinity binding sites (Kd = 3.1 ± 1.3 nM; 11.7 pmol/mg of membrane protein). This binding could be displaced by the μ-receptor selective agonist DAMGO (Fig. 3A), but not by either the δ-selective agonist [d-Pen2, d-Pen5]enkephalin (DPDPE; Fig. 3B) or the κ-selective agonist U50488 (Fig. 3C). Saturation binding with [3H]DAMGO revealed two classes of binding sites (Fig. 2B) with Kd values of 0.63 ± 0.15 nM (0.95 pmol/mg) and 3.8 ± 0.7 nM (2.55 pmol/mg). Added together, these two sites represent 3.5 pmol/mg, which is considerably less than the number of [3H]naloxone binding sites. These data indicate that not all of the naloxone binding sites have high affinity for DAMGO, and they are in agreement with the observation (Fig. 3A) that the displacement of [3H]naloxone binding by DAMGO is incomplete. A plausible explanation for these findings (Figs. 2 and 3) is that the expression of a receptor, that derives from a fish that lives in cold temperatures, in a human cell-line, results in a substantial proportion of receptors having an incorrect agonist-binding conformation at 37°C.
Figure 2.
Binding of opioid receptor ligands. Saturation binding experiments, with either the nonselective opioid receptor antagonist [3H]naloxone (A) or the μ-selective agonist [3H]DAMGO (B) to membranes prepared from HEK 293 cells expressing the C. commersoni μ-opioid receptor, were carried out as described. Specific binding was calculated by subtracting the nonspecific binding (determined in the presence of 10 μM naloxone) from the total binding determined using a range of concentrations of either [N-allyl-2,3-3H]naloxone (0.1–30 nM) or [3H]DAMGO (0.1–30 nM). (Insets) Scatchard plots calculated from the binding data shown.
Figure 3.
Displacement of [3H]naloxone binding by DAMGO but not by either DPDPE or U50488. The binding of 10 nM [N-allyl-2,3-3H]naloxone to membranes prepared from HEK 293 cells expressing the C. commersoni μ-opioid receptor was determined in the presence of a range of concentrations of the μ-selective agonist DAMGO (0.1 nM to 10 μM; A), the δ-selective agonist DPDPE (0.1 nM to 10 μM; B), and the κ-selective agonist U50488 (0.1 nM to 3 μM; C).
Functional Coupling of the Teleost Fish μ-Opioid Receptor.
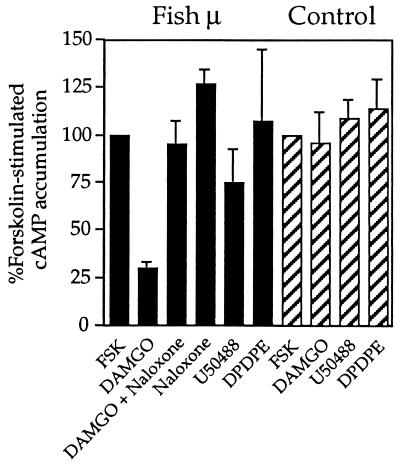
To study the effect of activation of the fish μ-opioid receptor, we measured the effect of 1 μM DAMGO on forskolin-stimulated cAMP accumulation in transfected HEK 293 cells. This agonist decreased the amount of cAMP by 74 ± 3%, and 10 μM naloxone completely abolished this effect (Fig. 4). In contrast, 1 μM U50488 caused only a slight reduction (25 ± 18%) in cAMP levels, and 1 μM DPDPE had no effect. Interestingly, naloxone treatment alone resulted in a slight increase in cAMP levels. This suggests that some spontaneous activation of the receptor, which can be blocked by the antagonist, occurs in HEK 293 cells. Lastly, none of the agonists had an effect on forskolin-stimulated cAMP accumulation in untransfected cells (Fig. 4), which is in agreement with the absence of [3H]naloxone binding sites (data not shown).
Figure 4.
Functional negative coupling to adenylate cyclase. Subtype-selective opioid receptor agonists were tested for their ability to inhibit forskolin-stimulated cAMP accumulation in HEK 293 cells expressing the C. commersoni μ-opioid receptor (filled bars) and in nontransfected cells (striped bars). Cells were incubated in the presence of 16 nM forskolin (FSK) alone or together with either DAMGO (1 μM), DAMGO (1 μM) plus naloxone (10 μM), naloxone (10 μM), U50488 (1 μM), or DPDPE (1 μM).
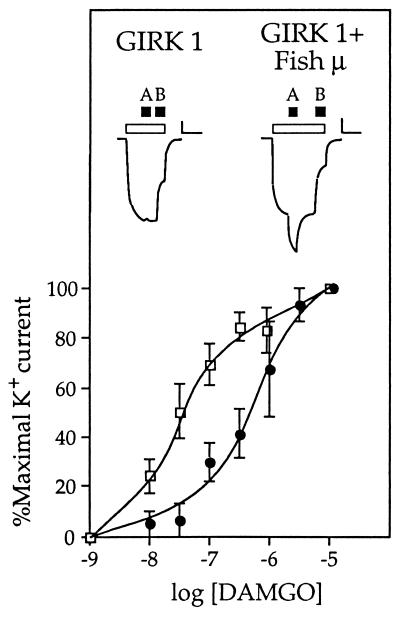
Xenopus oocytes that had been injected with in vitro-transcribed RNA for the C. commersoni μ-opioid receptor did not respond to the application of any opioid receptor ligand (data not shown). This indicates that the receptor is not coupled, via G-proteins of the Gq/G11 family, to the activation of phospholipase C. It has previously been shown that agonist binding to opioid receptors can result in the activation of an inward-rectifying potassium channel (26). The gating of this channel is due to a direct interaction with free βγ subunits of heterotrimeric G-proteins (27). After injection of Xenopus oocytes with in vitro-transcribed RNA for the mouse GIRK1, no response was observed upon the application of opioid receptor ligands (Fig. 5). However, coinjection of RNA for the fish μ-receptor and GIRK1 resulted in potassium channels that could be efficiently and dose-dependently activated by DAMGO (Fig. 5), but not by U50488 or DPDPE. A comparison of the coupling to GIRK1, of the fish and rat μ-receptors, revealed that the EC50 value for activation of the mammalian receptor is 10-fold lower (0.04 μM) than that (0.4 μM) of the piscine receptor. The difference in the EC50 values presumably reflects differences in the coupling efficiencies between the mouse channel and the fish and rat receptors.
Figure 5.
Functional coupling to mouse GIRK1. Dose-response curves are shown for the DAMGO-induced peak currents obtained from Xenopus oocytes expressing GIRK1 and either the C. commersoni μ-opioid receptor (•) or the rat μ-opioid receptor (□). Above these are displayed typical current traces recorded from an oocyte expressing only GIRK1 (Left) and from an oocyte expressing GIRK1 and the fish μ-opioid receptor (Right). Open bars indicate the period of time during which the perfusion medium was changed from ND-96 to hK; the solid bars denote the subsequent application of 1 μM DAMGO (bar A) and 300 μM BaCl2 (bar B). Note that in oocytes expressing either GIRK1 alone or GIRK1 and a μ-opioid receptor, changing from a bathing medium with a low potassium concentration (2 mM; ND-96) to one with a high potassium concentration (96 mM; hK) induces an inward current due to endogenous potassium channels and basal activation of GIRK1. A subsequent response to DAMGO is only seen when a μ-opioid receptor is present. The application of BaCl2, which specifically blocks GIRK1 currents, permits determination of the contribution of this channel to the total current response. (Bars: vertical, 60 nA; horizontal, 120 sec)
Tissue Distribution of the Teleost Fish μ-Opioid Receptor.
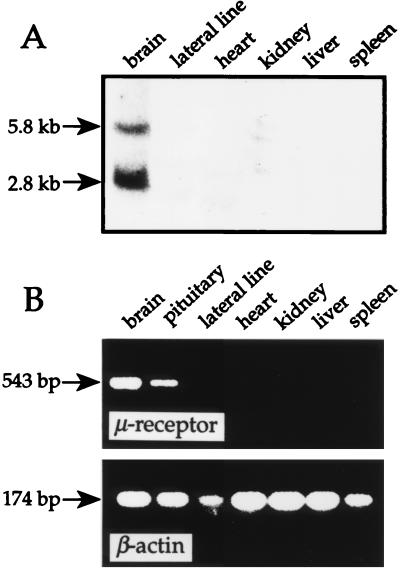
As assessed by Northern blot analysis of total RNA, the μ-opioid receptor gene is expressed in the teleost fish brain (Fig. 6A); there, two transcripts of 2.8 and 5.8 kb are detected. Due to a shortage of tissue, it was not possible to include pituitary RNA on the Northern blot. However, RT-PCR (35 cycles of amplification) revealed that the fish μ-opioid receptor gene is expressed in this tissue (Fig. 6B). At higher cycle number, very weak expression was also observed in liver (data not shown). The RT-PCR amplifications with β-actin primers (Fig. 6B) confirmed the integrity of the cDNAs used.
Figure 6.
Tissue expression of the C. commersoni μ-opioid receptor gene. Shown are the results of a Northern blot analysis (A) and an RT-PCR experiment (B). The numbers on the left indicate the estimated transcript sizes (A) and the amplified cDNA fragment sizes (B). For details, see Materials and Methods.
DISCUSSION
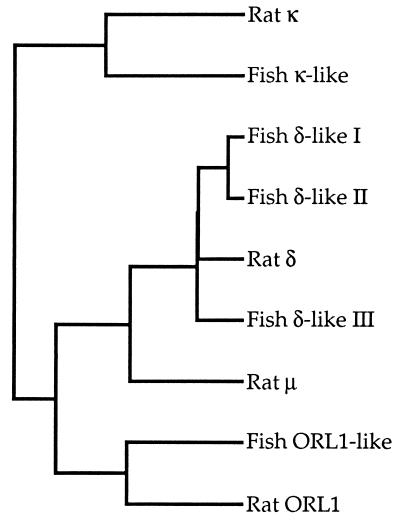
To date, in mammals, four opioid receptors have been identified in cDNA cloning studies. From the lower vertebrate C. commersoni, a teleost fish, we have isolated a full-length cDNA for a μ-receptor plus five partial clones that encode distinct opioid receptor-like proteins (Fig. 7); of the latter, one resembles the κ-receptor, one is similar to the ORL1 receptor, and three appear related to the δ-receptor. When compared with the latest update of the EMBL/GenBank database, all of the partial sequences are seen to most closely resemble mammalian opioid receptors. Our results can be interpreted in one of two ways: either the number of distinct opioid receptors has decreased during the evolution of vertebrates or there exist additional mammalian opioid receptors for which cDNAs have yet to be isolated. Evidence for the latter is suggested by pharmacological experiments that have distinguished subtypes of the different mammalian opioid receptors (3).
Figure 7.
A family of opioid receptor-like sequences exists in C. commersoni. The dendrogram shows the relationships of the partial sequences of five teleost fish opioid-like receptors to the rat μ- (11), κ- (9) and δ- (11) opioid receptors, and the rat ORL1 receptor (14); this was generated using the computer program pileup (Wisconsin Sequence Analysis Package).
When the teleost fish μ-opioid receptor is expressed in HEK 293 cells, we observe a single class of high-affinity binding sites for [3H]naloxone. However, saturation binding experiments, to the same membranes, revealed two classes of binding sites for [3H]DAMGO (Kd = 0.63 ± 0.15 nM and 3.8 ± 0.7 nM). The site with a Kd value of 0.63 nM probably corresponds to that population of receptors which is functionally coupled to a G-protein; this affinity is comparable to the Kd of 0.4 nM which has been reported for the rat μ-opioid receptor expressed in COS cells (13). The site with a lower affinity for [3H]DAMGO likely represents receptors that are not coupled to a G-protein. Note that two types of DAMGO binding sites have previously been reported for the expressed rat μ-opioid receptor (28).
We have shown that the fish μ-opioid receptor can functionally couple to G-protein-linked signaling pathways. Thus, DAMGO substantially reduced forskolin-stimulated cAMP levels in receptor-expressing HEK 293 cells. This is in line with the inhibitory effect on adenylate cyclase that has been observed with all opioid receptors identified to date (18). In Xenopus oocytes, strong activation of a coexpressed G-protein-gated potassium channel was produced by the application of DAMGO, a feature that has also been described for the rat μ-receptor (26). This is, perhaps, not surprising given the high degree of sequence identity seen, in the third intracellular loop and the region just C-terminal of TM7, between the fish and rat μ-opioid receptors. These two regions have been shown to be important in determining the specificity of G-protein coupling in other members of the G-protein-coupled receptor superfamily (29).
In conclusion, our data indicate that a diversity of opioid receptors was present early in vertebrate evolution, and that the structural and functional properties of the μ-receptor have been highly conserved over a period of ≈400 million years, which is the evolutionary distance between teleost fish and modern mammals (30). This is supported by observations of enkephalin-containing and β-endorphin-containing neurons in the brains of several species of fish (31, 32). Thus, the μ-opioid receptor arose very early in evolution, perhaps before the appearance of vertebrates. Although, as far as we are aware, an opioid receptor has not yet been cloned from any invertebrate, there is ample evidence in support of the existence of opioid peptides in lower animals such as molluscs (ref. 33, and references cited therein). If the function of opioid receptors in mammals is to suppress pain, then either they fulfill the same role in lower vertebrates, or they have evolved an antinociceptive function during the course of evolution.
Acknowledgments
We thank Dirk van den Boom and Christian Jurinke for help during the initial stages of this work, Dr. Volker Höllt (Magdeburg) for the mouse GIRK1 and rat μ-opioid receptor cDNAs, Thomas Kroslak (Magdeburg) for help with the initial oocyte recordings, Hans-Hinrich Hönck for membrane preparation and oocyte injection, and Helen Wilson (Nanton, Alberta, Canada) for help with the collection of C. commersoni. The data presented here form part of a thesis by F.R.G.
ABBREVIATIONS
- DAMGO
[d-Ala2, MePhe4-Gly-ol5]enkephalin
- DPDPE
[d-Pen2, d-Pen5]enkephalin
- RT-PCR
reverse transcription–PCR
- GIRK1
G-protein-gated inward-rectifying potassium channel.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y10904).
References
- 1.Goldstein A. Trends Pharmacol Sci. 1987;8:456–459. [Google Scholar]
- 2.Loh H H, Smith A P. Annu Rev Pharmacol Toxicol. 1990;30:123–147. doi: 10.1146/annurev.pa.30.040190.001011. [DOI] [PubMed] [Google Scholar]
- 3.Barnard E A, Simon J. In: Neurotransmitter Receptors. Hucho F, editor. Amsterdam: Elsevier; 1993. pp. 297–323. [Google Scholar]
- 4.Evans C J, Keith D E, Jr, Morrison H, Magendzo K, Edwards R H. Science. 1992;258:1952–1955. doi: 10.1126/science.1335167. [DOI] [PubMed] [Google Scholar]
- 5.Kieffer B L, Befort K, Gaveriaux-Ruff C, Hirth C G. Proc Natl Acad Sci USA. 1992;89:12048–12052. doi: 10.1073/pnas.89.24.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasuda K, Raynor K, Kong H, Breder C D, Takeda J, Reisine T, Bell G I. Proc Natl Acad Sci USA. 1993;90:6736–6740. doi: 10.1073/pnas.90.14.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng F, Xie G-X, Thompson R C, Mansour A, Goldstein A, Watson S J, Akil H. Proc Natl Acad Sci USA. 1993;90:9954–9958. doi: 10.1073/pnas.90.21.9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minami M, Toya T, Katao Y, Maekawa K, Nakamura S, Onogi T, Kaneko S, Satoh M. FEBS Lett. 1993;329:291–295. doi: 10.1016/0014-5793(93)80240-u. [DOI] [PubMed] [Google Scholar]
- 9.Nishi M, Takeshima H, Fukuda K, Kato S, Mori K. FEBS Lett. 1993;330:77–80. doi: 10.1016/0014-5793(93)80923-i. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Mestek A, Liu J, Hurley J A, Yu L. Mol Pharmacol. 1993;44:8–12. [PubMed] [Google Scholar]
- 11.Fukuda K, Kato S, Mori K, Nishi M, Takeshima H. FEBS Lett. 1993;327:311–314. doi: 10.1016/0014-5793(93)81011-n. [DOI] [PubMed] [Google Scholar]
- 12.Thompson R C, Mansour A, Akil H, Watson S J. Neuron. 1993;11:903–913. doi: 10.1016/0896-6273(93)90120-g. [DOI] [PubMed] [Google Scholar]
- 13.Wang J B, Imai Y, Eppler C M, Gregor P, Spivak C E, Uhl G R. Proc Natl Acad Sci USA. 1993;90:10230–10234. doi: 10.1073/pnas.90.21.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda K, Kato S, Mori K, Nishi M, Takeshima H, Iwabe N, Miyata T, Houtani T, Sugimoto T. FEBS Lett. 1994;343:42–46. doi: 10.1016/0014-5793(94)80603-9. [DOI] [PubMed] [Google Scholar]
- 15.Mollereau C, Parmentier M, Mailleux P, Butour J-L, Moisand C, Chalon P, Caput D, Vassart G, Meunier J-C. FEBS Lett. 1994;341:33–38. doi: 10.1016/0014-5793(94)80235-1. [DOI] [PubMed] [Google Scholar]
- 16.Meunier J-C, Mollereau C, Toll L, Suaudeau C, Moisand C, Alvinerie P, Butour J-L, Guillemot J-C, Ferrara P, Monsarrat B, Mazarguil H, Vassart G, Parmentier M, Costentin J. Nature (London) 1995;377:532–535. doi: 10.1038/377532a0. [DOI] [PubMed] [Google Scholar]
- 17.Reinscheid R K, Nothacker H-P, Bourson A, Ardati A, Henningsen R A, Bunzow J R, Grandy D K, Langen H, Monsma F J, Jr, Civelli O. Science. 1995;270:792–794. doi: 10.1126/science.270.5237.792. [DOI] [PubMed] [Google Scholar]
- 18.Reisine T, Bell G I. Trends Neurosci. 1993;16:506–510. doi: 10.1016/0166-2236(93)90194-q. [DOI] [PubMed] [Google Scholar]
- 19.Matthes H W D, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, Befort K, Dierich A, Le Meur M, Dollé P, Tzavara E, Hanoune J, Roques B P, Kieffer B L. Nature (London) 1996;383:819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 20.Iversen L L. Nature (London) 1996;383:759–760. doi: 10.1038/383759a0. [DOI] [PubMed] [Google Scholar]
- 21.Frohman M A, Martin G R. Technique. 1989;1:165–170. [Google Scholar]
- 22.Harvey R J, Vreugdenhil E, Zaman S H, Bhandal N S, Usherwood P N R, Barnard E A, Darlison M G. EMBO J. 1991;10:3239–3245. doi: 10.1002/j.1460-2075.1991.tb04887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stühmer T, Amar M, Harvey R J, Bermudez I, van Minnen J, Darlison M G. J Neurosci. 1996;16:2869–2880. doi: 10.1523/JNEUROSCI.16-09-02869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingston R E. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Greene; 1987. pp. 9.0.1–9.5.6. [Google Scholar]
- 25.Roth A, Kreienkamp H-J, Nehring R B, Roosterman D, Meyerhof W, Richter D. DNA Cell Biol. 1997;16:111–119. doi: 10.1089/dna.1997.16.111. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Yu L. J Biol Chem. 1994;269:7839–7842. [PubMed] [Google Scholar]
- 27.Reuveny E, Slesinger P A, Inglese J, Morales J M, Iñiguez-Lluhi J A, Lefkowitz R J, Bourne H R, Jan Y N, Jan L Y. Nature (London) 1994;370:143–146. doi: 10.1038/370143a0. [DOI] [PubMed] [Google Scholar]
- 28.Zastawny R L, George S R, Nguyen T, Cheng R, Tsatsos J, Briones- Urbina R, O’Dowd B F. J Neurochem. 1994;62:2099–2105. doi: 10.1046/j.1471-4159.1994.62062099.x. [DOI] [PubMed] [Google Scholar]
- 29.Baldwin J M. Curr Opin Cell Biol. 1994;6:180–190. doi: 10.1016/0955-0674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 30.Kardong K V. Vertebrates: Comparative Anatomy, Function, Evolution. Dubuque, IA: Brown; 1995. [Google Scholar]
- 31.Stuesse S L, Stuesse D C, Cruce W L. J Comp Neurol. 1995;358:414–427. doi: 10.1002/cne.903580308. [DOI] [PubMed] [Google Scholar]
- 32.Vallarino M, Delbende C, Bunel D T, Ottonello I, Vaudry H. Peptides. 1989;10:1223–1230. doi: 10.1016/0196-9781(89)90016-8. [DOI] [PubMed] [Google Scholar]
- 33.Carpenter D O, Kemenes G, Elekes K, Leung M, Stefano G, S-, Rózsa K, Salánki J. Cell Mol Neurobiol. 1995;15:239–256. doi: 10.1007/BF02073331. [DOI] [PubMed] [Google Scholar]