Abstract
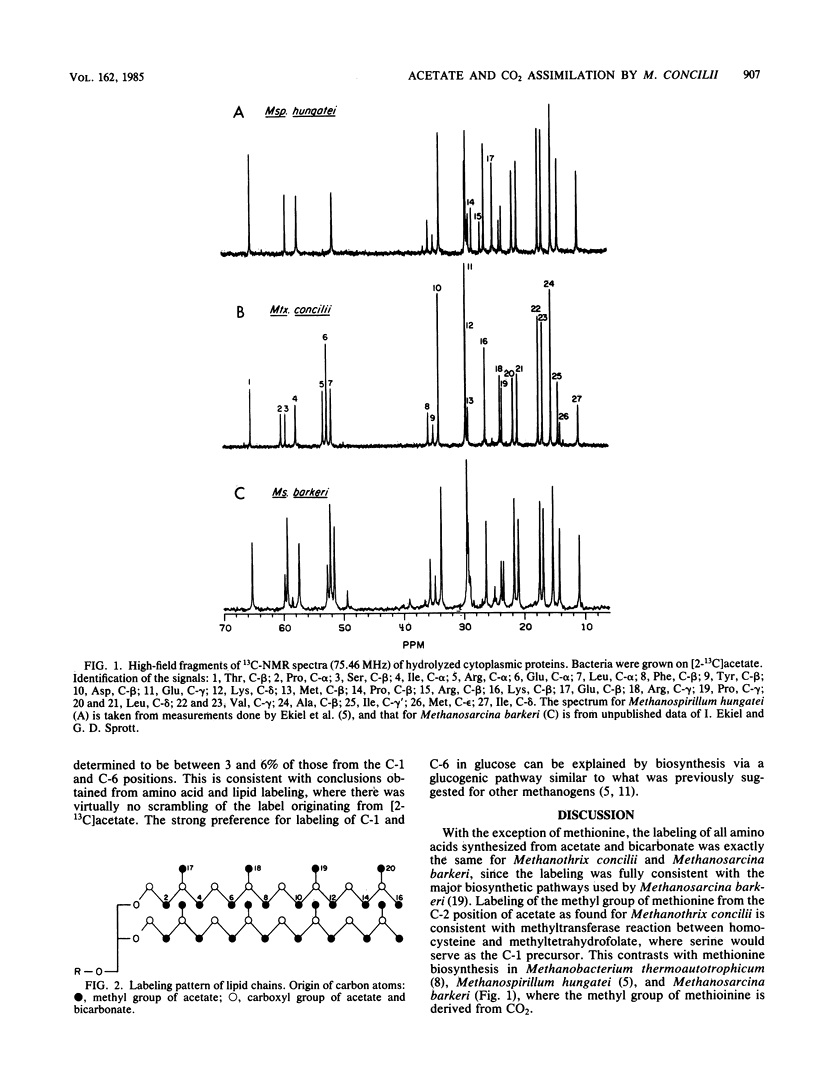
Biosynthetic pathways in Methanothrix concilii, a recently isolated aceticlastic methanogen, were studied by 13C-nuclear magnetic resonance spectroscopy. Labeling patterns of amino acids, lipids, and carbohydrates were determined. Similar to other methanogens, acetate was carboxylated to pyruvate, which was further converted to amino acids by various biosynthetic pathways. The origin of carbon atoms in glutamate, proline, and arginine clearly showed that an incomplete tricarboxylic acid cycle operating in the oxidative direction was used for their biosynthesis. Isoleucine was synthesized via citramalate, which is a typical route for methanogens. As with Methanosarcina barkeri, an extensive exchange of the label between the carboxyl group of acetate and CO2 was observed. Lipids predominantly contained diphytanyl chains, the labeling of which indicated that biosynthesis proceeded through mevalonic acid. Labeling of the C-1,6 of glucose from [2-13C]acetate is consistent with a glucogenic route for carbohydrate biosynthesis. Except for the different origins of the methyl group of methionine, the metabolic properties of Methanothrix concilii are closely related to those of Methanosarcina barkeri.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Degani H., Danon A., Caplan S. R. Proton and carbon-13 nuclear magnetic resonance studies of the polar lipids of Halobacterium halobium. Biochemistry. 1980 Apr 15;19(8):1626–1631. doi: 10.1021/bi00549a016. [DOI] [PubMed] [Google Scholar]
- Ekiel I., Smith I. C., Sprott G. D. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J Bacteriol. 1983 Oct;156(1):316–326. doi: 10.1128/jb.156.1.316-326.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten T. J., Bongaerts H. C., van der Drift C., Vogels G. D. Acetate, methanol and carbon dioxide as substrates for growth of Methanosarcina barkeri. Antonie Van Leeuwenhoek. 1980;46(6):601–610. doi: 10.1007/BF00394016. [DOI] [PubMed] [Google Scholar]
- Mah R. A., Smith M. R., Baresi L. Studies on an acetate-fermenting strain of Methanosarcina. Appl Environ Microbiol. 1978 Jun;35(6):1174–1184. doi: 10.1128/aem.35.6.1174-1184.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. H., Mah R. A. Kinetics of acetate metabolism during sludge digestion. Appl Microbiol. 1966 May;14(3):368–371. doi: 10.1128/am.14.3.368-371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer P. J., Zeikus J. G. Acetate assimilation pathway of Methanosarcina barkeri. J Bacteriol. 1979 Jan;137(1):332–339. doi: 10.1128/jb.137.1.332-339.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehnder A. J., Huser B. A., Brock T. D., Wuhrmann K. Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch Microbiol. 1980 Jan;124(1):1–11. doi: 10.1007/BF00407022. [DOI] [PubMed] [Google Scholar]


