Abstract
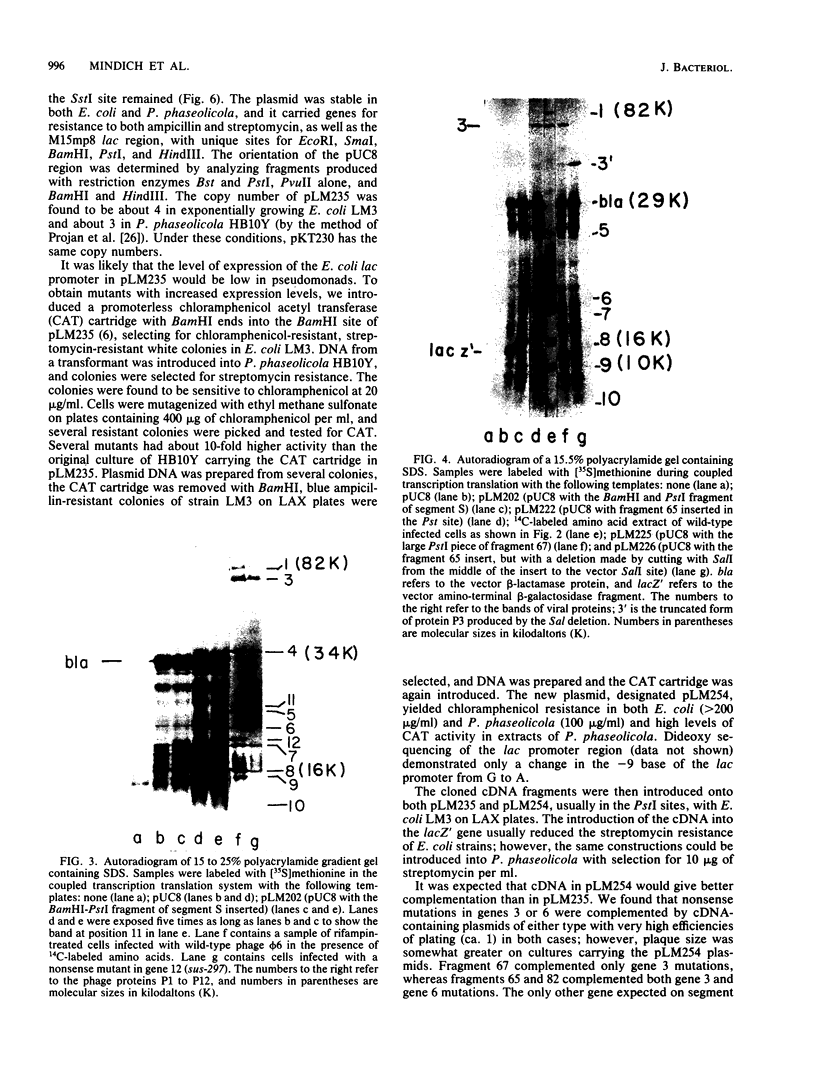
Phage phi 6 has a genome consisting of three pieces of double-stranded RNA. Single-stranded RNA was prepared from phi 6 nucleocapsids by in vitro transcription with the phage RNA polymerase. These transcripts were polyadenylated and used as templates for the preparation of cDNA copies. The resulting DNA was cloned into the PstI restriction nuclease site of plasmid pBR322. Insert-bearing plasmids were annealed to phi 6 RNA to assign the inserts to their proper segments. In this way we identified inserts corresponding to the large, medium, and small segments. Two large overlapping inserts of the small segment constitute the complete complement of the segment as determined by the sequence analysis of the DNA. In vitro coupled transcription and translation showed that the small segment inserts were able to direct the synthesis of the four known genes in the small segment. Two overlapping inserts in the medium segment constitute the entire segment and were shown to direct the in vitro synthesis of two of the three known proteins of the medium segment. Several inserts bearing about one-third the complement of the large segment were also isolated, and one of these directed the synthesis of a peptide that resembles protein P1. Restriction endonuclease maps were prepared for the inserts, and by in vitro synthesis it was possible to refine the genetic map of phi 6. A chimeric plasmid was constructed that combines plasmids pUC8 and RSF1010. Inserts placed on this plasmid were transformed to Pseudomonas phaseolicola, the natural host of phage phi 6. It was possible to refine further the genetic map by complementation of nonsense mutants of phi 6 with the cDNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bamford D. H., Mindich L. Electron microscopy of cells infected with nonsense mutants of bacteriophage phi 6. Virology. 1980 Nov;107(1):222–228. doi: 10.1016/0042-6822(80)90287-1. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Bothwell A. L., Paskind M., Reth M., Imanishi-Kari T., Rajewsky K., Baltimore D. Heavy chain variable region contribution to the NPb family of antibodies: somatic mutation evident in a gamma 2a variable region. Cell. 1981 Jun;24(3):625–637. doi: 10.1016/0092-8674(81)90089-1. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Day L. A., Mindich L. The molecular weight of bacteriophage phi 6 and its nucleocapsid. Virology. 1980 Jun;103(2):376–385. doi: 10.1016/0042-6822(80)90196-8. [DOI] [PubMed] [Google Scholar]
- Derman E., Krauter K., Walling L., Weinberger C., Ray M., Darnell J. E., Jr Transcriptional control in the production of liver-specific mRNAs. Cell. 1981 Mar;23(3):731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- Devos R., van Emmelo J., Seurinck-Opsomer C., Gillis E., Fiers W. Addition by ATP: RNA adenylyltransferase from Escherichia coli of 3'-linked oligo(A) to bacteriophage Qbeta RNA and its effect on RNA replication. Biochim Biophys Acta. 1976 Oct 18;447(3):319–327. doi: 10.1016/0005-2787(76)90055-1. [DOI] [PubMed] [Google Scholar]
- Gantotti B. V., Patil S. S., Mandel M. Genetic transformation of Pseudomonas phaseolicola by R-factor DNA. Mol Gen Genet. 1979 Jul 2;174(1):101–103. doi: 10.1007/BF00433310. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Iba H., Watanabe T., Emori Y., Okada Y. Three double-stranded RNA genome segments of bacteriophage phi 6 have homologous terminal sequences. FEBS Lett. 1982 May 3;141(1):111–115. doi: 10.1016/0014-5793(82)80027-6. [DOI] [PubMed] [Google Scholar]
- Kakitani H., Iba H., Okada Y. Penetration and partial uncoating of bacteriophage phi 6 particle. Virology. 1980 Mar;101(2):475–483. doi: 10.1016/0042-6822(80)90461-4. [DOI] [PubMed] [Google Scholar]
- Land H., Grez M., Hauser H., Lindenmaier W., Schütz G. 5'-Terminal sequences of eucaryotic mRNA can be cloned with high efficiency. Nucleic Acids Res. 1981 May 25;9(10):2251–2266. doi: 10.1093/nar/9.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman J. F., Mindich L. The isolation of new mutants of bacteriophage phi 6. Virology. 1979 Aug;97(1):164–170. doi: 10.1016/0042-6822(79)90382-9. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mindich L., Bamford D., Goldthwaite C., Laverty M., Mackenzie G. Isolation of nonsense mutants of lipid-containing bacteriophage PRD1. J Virol. 1982 Dec;44(3):1013–1020. doi: 10.1128/jvi.44.3.1013-1020.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L., Cohen J., Weisburd M. Isolation of nonsense suppressor mutants in Pseudomonas. J Bacteriol. 1976 Apr;126(1):177–182. doi: 10.1128/jb.126.1.177-182.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L., Lehman J. Characterization of phi 6 mutants that are temperature sensitive in the morphogenetic protein P12. Virology. 1983 Jun;127(2):438–445. doi: 10.1016/0042-6822(83)90156-3. [DOI] [PubMed] [Google Scholar]
- Mindich L., Lehman J., Huang R. Temperature-dependent compositional changes in the envelope of phi 6. Virology. 1979 Aug;97(1):171–176. doi: 10.1016/0042-6822(79)90383-0. [DOI] [PubMed] [Google Scholar]
- Mindich L., Sinclair J. F., Cohen J. The morphogenesis of bacteriophage phi6: particles formed by nonsense mutants. Virology. 1976 Nov;75(1):224–231. doi: 10.1016/0042-6822(76)90021-0. [DOI] [PubMed] [Google Scholar]
- Mindich L., Sinclair J. F., Levine D., Cohen J. Genetic studies of temperature-sensitive and nonsense mutants of bacteriophage phi6. Virology. 1976 Nov;75(1):218–223. doi: 10.1016/0042-6822(76)90020-9. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Carleton S., Novick R. P. Determination of plasmid copy number by fluorescence densitometry. Plasmid. 1983 Mar;9(2):182–190. doi: 10.1016/0147-619x(83)90019-7. [DOI] [PubMed] [Google Scholar]
- Roychoudhury R., Jay E., Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976 Jan;3(1):101–116. doi: 10.1093/nar/3.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semancik J. S., Vidaver A. K., Van Etten J. L. Characterization of segmented double-helical RNA from bacteriophage phi6. J Mol Biol. 1973 Aug 25;78(4):617–625. doi: 10.1016/0022-2836(73)90283-0. [DOI] [PubMed] [Google Scholar]
- Sinclair J. F., Cohen J., Mindich L. The isolation of suppressible nonsence mutants of bacteriophage phi6. Virology. 1976 Nov;75(1):198–208. [PubMed] [Google Scholar]
- Sinclair J. F., Tzagoloff A., Levine D., Mindich L. Proteins of bacteriophage phi6. J Virol. 1975 Sep;16(3):685–695. doi: 10.1128/jvi.16.3.685-695.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaver A. K., Koski R. K., Van Etten J. L. Bacteriophage phi6: a Lipid-Containing Virus of Pseudomonas phaseolicola. J Virol. 1973 May;11(5):799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Komaroff L., Efstratiadis A., Broome S., Lomedico P., Tizard R., Naber S. P., Chick W. L., Gilbert W. A bacterial clone synthesizing proinsulin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3727–3731. doi: 10.1073/pnas.75.8.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. L., Ivashkiv L., Chen H. Z., Zubay G., Cashel M. Cell-free coupled transcription-translation system for investigation of linear DNA segments. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7029–7033. doi: 10.1073/pnas.77.12.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]






