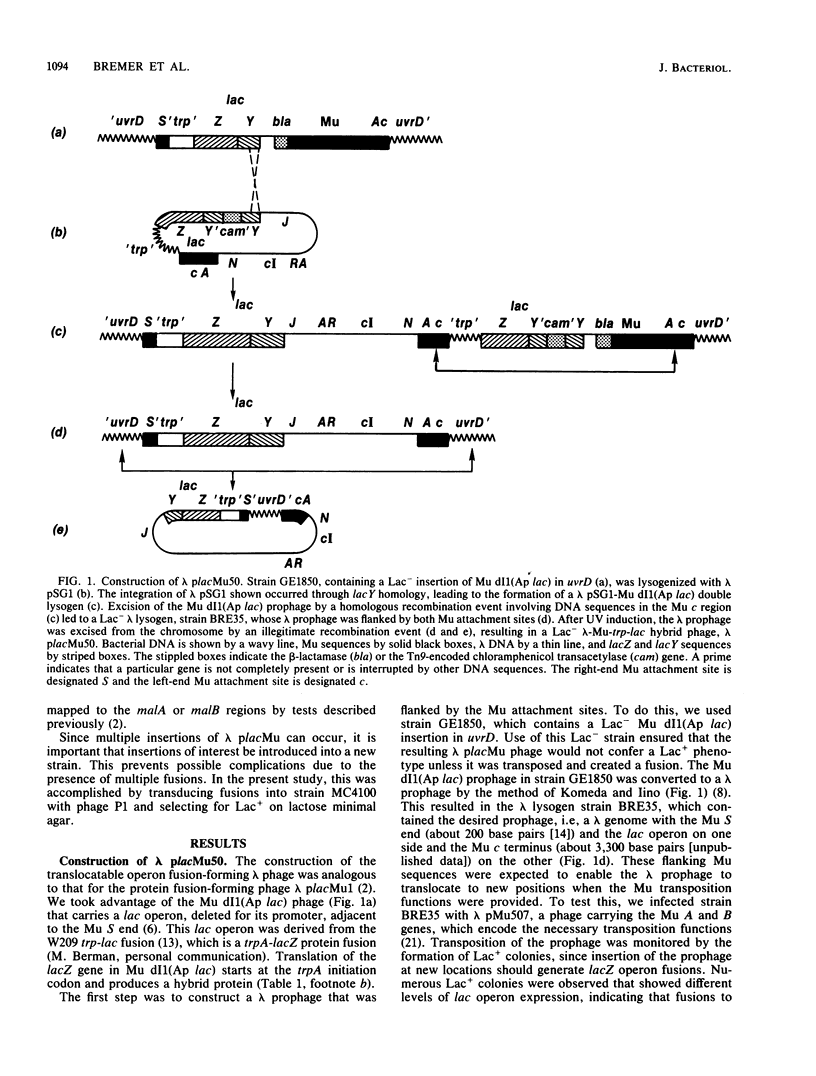
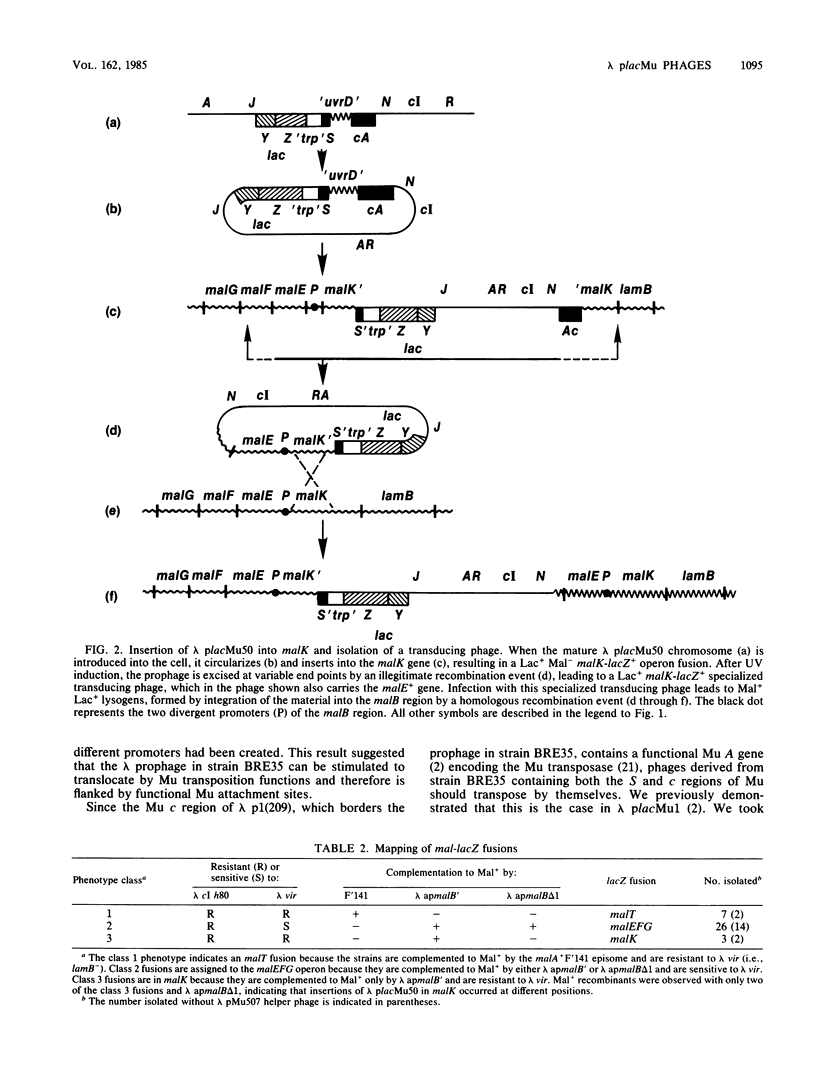
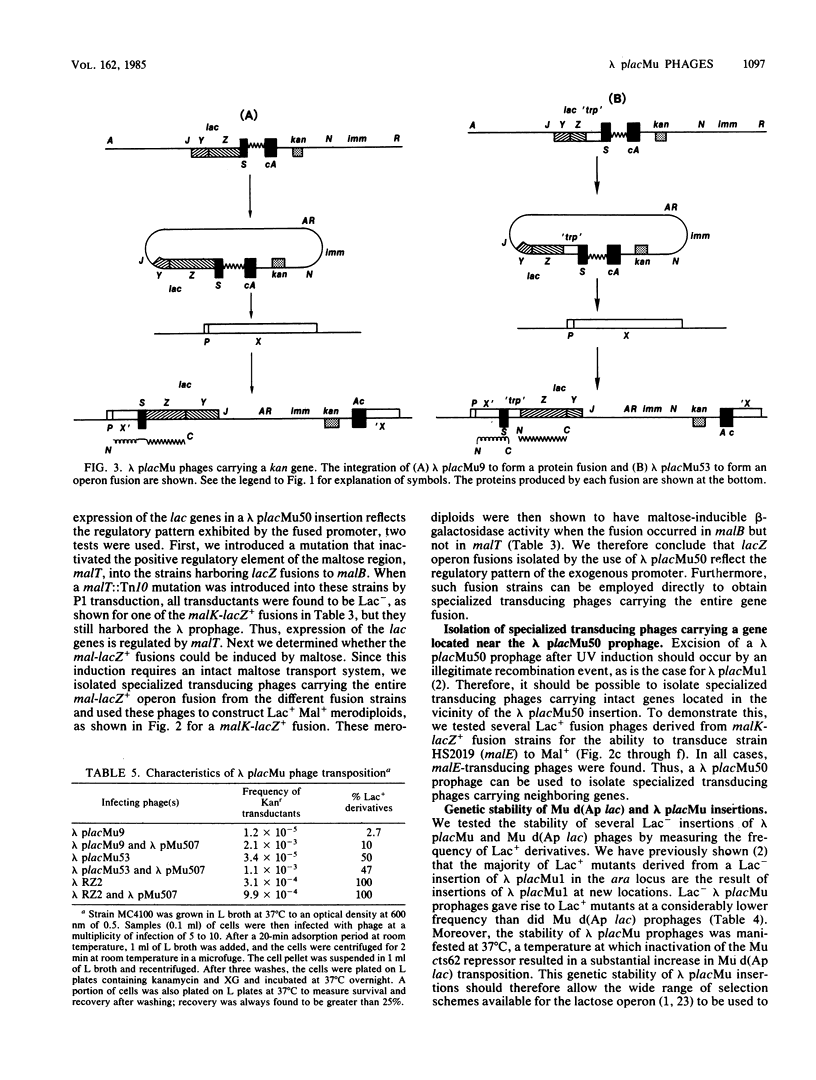
Abstract
We have constructed several derivatives of bacteriophage lambda that translocate by using the transposition machinery of phage Mu (lambda placMu phages). Each phage carries the c end of Mu, containing the Mu cIts62, ner (cII), and A genes, and the terminal sequences from the Mu S end (beta end). These sequences contain the Mu attachment sites, and their orientation allows the lambda genome to be inserted into other chromosomes, resulting in a lambda prophage flanked by the Mu c and S sequences. These phages provide a means to isolate cells containing fusions of the lac operon to other genes in vivo in a single step. In lambda placMu50, the lacZ and lacY genes, lacking a promoter, were located adjacent to the Mu S sequence. Insertion of lambda placMu50 into a gene in the proper orientation created an operon fusion in which lacZ and lacY were expressed from the promoter of the target gene. We also introduced a gene, kan, which confers kanamycin resistance, into lambda placMu50 and lambda placMu1, an analogous phage for constructing lacZ protein fusions (Bremer et al., J. Bacteriol. 158:1084-1093, 1984). The kan gene, located between the cIII and ssb genes of lambda, permitted cells containing insertions of these phages to be selected independently of their Lac phenotype.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bremer E., Silhavy T. J., Weisemann J. M., Weinstock G. M. Lambda placMu: a transposable derivative of bacteriophage lambda for creating lacZ protein fusions in a single step. J Bacteriol. 1984 Jun;158(3):1084–1093. doi: 10.1128/jb.158.3.1084-1093.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J. In vivo formation of gene fusions encoding hybrid beta-galactosidase proteins in one step with a transposable Mu-lac transducing phage. Proc Natl Acad Sci U S A. 1984 Jan;81(2):535–539. doi: 10.1073/pnas.81.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Fusion of the Escherichia coli lac genes to the ara promoter: a general technique using bacteriophage Mu-1 insertions. Proc Natl Acad Sci U S A. 1975 Mar;72(3):809–813. doi: 10.1073/pnas.72.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Kalnins A., Otto K., Rüther U., Müller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2(4):593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y., Iino T. Regulation of expression of the flagellin gene (hag) in Escherichia coli K-12: analysis of hag-lac gene fusions. J Bacteriol. 1979 Sep;139(3):721–729. doi: 10.1128/jb.139.3.721-729.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magazin M., Howe M., Allet B. Partial correlation of the genetic and physical maps of bacteriophage Mu. Virology. 1977 Apr;77(2):677–688. doi: 10.1016/0042-6822(77)90491-3. [DOI] [PubMed] [Google Scholar]
- Marchal C., Greenblatt J., Hofnung M. malB region in Escherichia coli K-12: specialized transducing bacteriophages and first restriction map. J Bacteriol. 1978 Dec;136(3):1109–1119. doi: 10.1128/jb.136.3.1109-1119.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. H., Reznikoff W. S., Beckwith J. R. Genetic fusions defining trp and lac operon regulatory elements. J Mol Biol. 1975 Apr 15;93(3):331–350. doi: 10.1016/0022-2836(75)90281-8. [DOI] [PubMed] [Google Scholar]
- O'Connor M. B., Malamy M. H. A new insertion sequence, IS121, is found on the Mu dI1 (Ap lac) bacteriophage and the Escherichia coli K-12 chromosome. J Bacteriol. 1983 Nov;156(2):669–679. doi: 10.1128/jb.156.2.669-679.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Clément J. M., Hofnung M. Structure of the malB region in Escherichia coli K12. III. Correlation of the genetic map with the restriction map. Mol Gen Genet. 1979 Jul 24;174(3):261–267. doi: 10.1007/BF00267798. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Shuman H. A. Active transport of maltose in Escherichia coli K12. Role of the periplasmic maltose-binding protein and evidence for a substrate recognition site in the cytoplasmic membrane. J Biol Chem. 1982 May 25;257(10):5455–5461. [PubMed] [Google Scholar]
- Silhavy T. J., Benson S. A., Emr S. D. Mechanisms of protein localization. Microbiol Rev. 1983 Sep;47(3):313–344. doi: 10.1128/mr.47.3.313-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Platt T., Crawford I. P., Nichols B. P., Christie G. E., Horowitz H., VanCleemput M., Wu A. M. The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res. 1981 Dec 21;9(24):6647–6668. doi: 10.1093/nar/9.24.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]


