Abstract
We have discovered a mutant loblolly pine (Pinus taeda L.) in which expression of the gene encoding cinnamyl alcohol dehydrogenase (CAD; EC 1.1.1.195) is severely reduced. The products of CAD, cinnamyl alcohols, are the precursors of lignin, a major cell wall polymer of plant vascular tissues. Lignin composition in this mutant shows dramatic modifications, including increased incorporation of the substrate of CAD (coniferaldehyde), indicating that CAD may modulate lignin composition in pine. The recessive cad-n1 allele, which causes this phenotype, was discovered in a tree heterozygous for this mutant allele. It is inherited as a simple Mendelian locus that maps to the same genomic region as the cad locus. In mutant plants, CAD activity and abundance of cad RNA transcript are low, and free CAD substrate accumulates to a high level. The wood of the mutant is brown, whereas the wood in wild types is nearly white. The wood phenotype resembles that of brown midrib (bm) mutants and some transgenic plants in which xylem is red-brown due to a reduction in CAD activity. However, unlike transgenics with reduced CAD, the pine mutant has decreased lignin content. Wood in which the composition of lignin varies beyond previous expectations still provides vascular function and mechanical support.
Lignin is a major component of the cell wall embedding matrix that reinforces the cellulose microfibrils of plant vascular tissues (1, 2). Thus, lignin confers compressive strength and bending stiffness necessary for mechanical support; it also provides a hydrophobic surface, essential for longitudinal water transport, and provides a barrier against pathogens. Lignin is a complex phenolic polymer, usually derived from various proportions of three cinnamyl alcohol‡‡ precursors, p-coumaryl, coniferyl, and sinapyl alcohols (3). The biological role of this variation in composition and its significance in the structure–function relationship of lignins remain poorly understood (4).
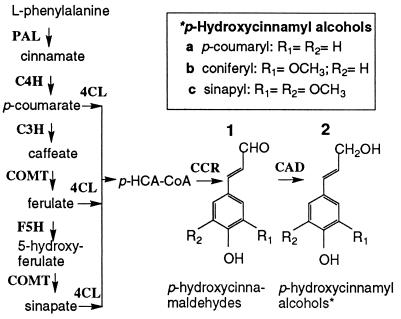
The precursors of lignin (monolignols) are products of a specialized branch of phenylpropanoid metabolism (Fig. 1). Many enzymes implicated in monolignol biosynthesis have been characterized and the corresponding genes have been cloned (1, 2). The last step of the monolignol pathway, the reduction of cinnamaldehydes to cinnamyl alcohols, is catalyzed by cinnamyl alcohol dehydrogenase (CAD; EC 1.1.1.195). Antisense suppression of CAD to as low as 7% of wild type in tobacco significantly increased the proportion of cinnamaldehyde subunits in lignin, but no effect was detected on the amount of lignin (5). The requirement for CAD in the synthesis of lignin precursors and the role of cinnamaldehydes as lignin precursors therefore need to be reassessed by more specific loss-of-function/gain-of-function experiments.
Figure 1.
Schematic of monolignol biosynthesis. The general phenylpropanoid pathway synthesizes the p-hydroxycinnamic acids (p-HCA) via the enzymes PAL (phenylalanine ammonia-lyase), C4H (cinnamate 4-hydroxylase), C3H (p-coumarate 3-hydroxylase), COMT (caffeate/5-hydroxyferulate O-methyltransferase), and F5H (ferulate 5-hydroxylase). Hydroxycinnamoyl-CoA thioesters (p-HCA-CoA), formed by 4CL (4 coumarate:CoA ligase) are reduced by CCR (cinnamoyl-CoA reductase) and CAD (cinnamyl alcohol dehydrogenase) to give p-hydroxycinnamyl alcohols.
In trees, lignin can account for 18–36% of the dry weight of wood (6). Therefore lignin mutants in tree species would be of particular interest to verify the biological role of lignin biosynthetic enzymes. However, lignin mutants have been reported only in herbaceous plants. Brown midrib (bm) mutants, found in monocots, have decreased lignin content and/or modified lignin composition (7, 8). Here, we identify and describe a cad mutant in a woody gymnosperm, loblolly pine (Pinus taeda L.). A mutant cad allele was identified in a heterozygous parent and genetically mapped in a haploid mapping population. This allele results in severely decreased cad gene expression and, in homozygotes, modifies wood color and lignin composition. This mutant provides new insights into the role of CAD and the biosynthesis of lignin in woody plants.
MATERIALS AND METHODS
Plant Material and Wood Sample Preparation.
Loblolly pine seeds, tissue, and wood samples were from the North Carolina State University Industry Cooperative Tree Improvement Program, Westvaco Corporation, and the Baruch Experimental Forest and were isolated as previously (9). Wood samples were debarked and dried at room temperature for 2 days [for milled wood lignin (MWL)] or at 65°C overnight and ground in a Wiley mill through a 60-mesh screen (10). Extractive free wood cell wall residue (CWR) was prepared according to Halpin et al. (5).
Allozyme Gels and Enzyme Assays.
Allozyme gel analyses used standard conditions for CAD activity (9, 11) and for glucose-6-phosphate dehydrogenase (G6PDH), alcohol dehydrogenase (ADH), and shikimic acid dehydrogenase (SDH) (12).
The reverse reaction of CAD (the oxidation of coniferyl alcohol to coniferaldehyde) was assayed (11) in whole-cell extracts (13) from 15–25 mg of fresh tissue per assay. Mean enzyme activities were obtained from six plants, each sampled in duplicate. The forward reaction (the reduction of coniferaldehyde to coniferyl alcohol) was assayed in partially purified protein extracts (11) from 150 mg of tissue, precipitated in the 30–70% range of saturated ammonium sulfate (14).
DNA Isolation, Random Amplified Polymorphic DNA (RAPD) Marker Analysis, and Genetic Mapping.
Genomic DNA was isolated from needles (15) and individual megagametophytes (Puregene DNA isolation kit, Gentra Systems). Approximately one-third of each haploid megagametophyte used in mapping experiments (5 to 6 mg) was used for allozyme gel analysis and the rest (9–12 mg) was stored at −80°C for DNA isolation. Methods for linkage analysis using RAPD markers and mapmaker for Macintosh version 2.0 (DuPont) were previously described (16, 17). For bulked segregant analysis (18), two pooled DNA samples each prepared from 12 megagametophytes and representing the two CAD allozymes of clone 6-1031 were screened with 170 RAPD primers (Operon Technologies, Alameda, CA) to identify linked markers. Cosegregation of RAPD markers and CAD allozymes was verified using 74 megagametophytes from clone 6-1031 and 108 megagametophytes from clone 7-56.
Isolation of RNA and Analysis of Transcript Levels.
Total RNA (19) for Northern blots was separated in 1.2% agarose gels with 2.2 M formaldehyde and transferred to Zetaprobe nylon membranes (Bio-Rad). A full-length cad cDNA (9) and a 170-bp fragment of a pine ribosomal protein cDNA were 32P radiolabeled with Ready-To-Go Labelling Beads (Pharmacia Biotech) and used as probes. The prehybridization, hybridization (in the presence of 50% deionized formamide), and membrane washes followed Brown and MacKey (20). Densitometric scanning of autoradiographs used a personal densitometer SI (Molecular Devices) and quantitations used the program ImageQuant (Molecular Devices).
Isolation and Analysis of Phenolic Extractives.
Phenolic extractives were recovered by successive extraction of ground wood with 50% ethanol (once) and 50% methanol (five times) at 70°C, each for 2 hr. Total phenolic content was determined with the Folin and Ciocalteu reagent (21), using ferulic acid as a standard. Extracts were hydrolyzed in 1 M NaOH for 1 hr at room temperature and analyzed by reverse-phase HPLC using a Microsorb C18 column (Rainin Instruments) with a 0% to 99% acetonitrile gradient in 1% acetic acid (21). Compounds were identified on the basis of coelution with standards and the UV spectrum of corresponding eluted fractions.
Isolation of Lignin and Fourier-Transform Infrared (FTIR) Analysis of Lignin and CWR.
MWL was prepared from one homozygous mutant and one wild-type 12-year old tree. Ground wood samples were ball milled (22) and extracted twice with 96:4 (vol/vol) dioxane/water (23). Extracts were freeze-dried, dissolved in 90% acetic acid, precipitated and washed with water and freeze-dried, dissolved in 2:1 (vol/vol) 1,2-dichloroethane/ethanol, precipitated with diethyl ether, washed with petroleum ether, and dried in vacuo. The yields of the MWL preparations were 19% of the total lignin for the mutant and 20% for the wild type.
FTIR transmission spectra of MWL and CWR were acquired using a Perkin–Elmer 16PC spectrophotometer. MWL (1 mg) or CWR (3 mg) was pressed into pellets with 300 mg of KBr. Spectral data were converted to absorbance units and vector-normalized using pecss software (Perkin–Elmer).
Lignin Content Determinations.
Lignin content of CWR was estimated by using both Klason and acetyl bromide (AcBr) methods. Klason acid-insoluble lignin was determined according to Effland (24), using samples of 180–200 mg of CWR. The A205 of the Klason acid filtrate was used to calculate the acid-soluble lignin (25). The AcBr method (26) used samples of 5–6 mg of CWR with or without a saponification pretreatment, consisting of three extractions with 1 M NaOH.
RESULTS
Detection and Inheritance of a Mutant cad Allele on Allozyme Gels.
In loblolly pine, CAD is encoded by a single gene which has several alleles encoding proteins of different electrophoretic mobilities (9). In many conifers, including pines, allelic variation is easily analyzed in the megagametophyte, a haploid (1n) seed tissue derived from the megaspore (27). Megagametophytes from a single tree can be used as a segregating haploid population, in which alleles at each locus heterozygous in the parent will segregate 1:1. Furthermore, the allele present in the megagametophyte is identical to the maternally inherited allele of the embryo in the same seed.
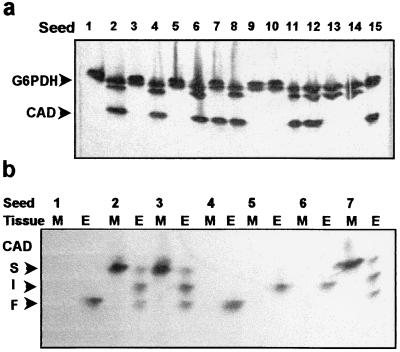
We uncovered a low-activity CAD allozyme segregating in megagametophytes from a heterozygous parent. In a genotype of loblolly pine (clone 7-56), megagametophytes from 50% of the seeds had a null allozyme phenotype for CAD (Fig. 2a), although all showed activity for G6PDH (Fig. 2a), shikimic acid dehydrogenase, and alcohol dehydrogenase (not shown). The segregation ratio of the low CAD activity phenotype was determined from 121 megagametophytes and was 62:59, not significantly different from a 1:1 ratio (χ2 = 0.074). These results are strong evidence that clone 7-56 is heterozygous for a mutation in a single genetic locus affecting CAD enzyme activity. This segregation ratio could result from a mutation with either a cis-acting or trans-acting effect on the cad gene product. To distinguish between these alternatives, the inheritance of the low-activity allozyme was analyzed in diploid tissue.
Figure 2.
Inheritance of the low-activity CAD allozyme. (a) Each lane contains megagametophyte extract from one seed of clone 7-56, costained for CAD and G6PDH. (b) Inheritance in megagametophytes (M) and embryos (E) within individual seeds, from a cross of two trees with four different cad alleles: the low-activity allozyme and a slow (S) mobility allozyme, clone 7-56, maternal parent; intermediate (I) and fast (F) alleles, from the pollen parent.
Inheritance patterns in the megagametophyte and embryo of individual seeds showed that offspring that inherit the low-activity allozyme from one parent (heterozygous offspring) have one active Cad allele. CAD is a dimeric protein, therefore, heterozygotes for fast and slow electrophoretic alleles show three bands for CAD in diploid tissues, consisting of two homodimers and one heterodimer (9). In controlled crosses of clone 7-56, maternal inheritance of the low-activity allozyme was unambiguously determined in megagametophytes and gave a single band of CAD activity in the embryos, a homodimeric band of staining contributed by the other parent (Fig. 2b). This type of staining pattern was observed in heterozygotes that inherited this allozyme paternally or maternally and was consistent with a single active Cad allele. The simplest explanation for these results is the inheritance of a cis-acting mutation that results in a mutant cad allele, designated cad-n1. By contrast, a dominant mutation in a trans-acting gene would suppress both Cad alleles, and a recessive trans-acting mutation would result in two active Cad alleles.
Genetic Mapping of the cad-n1 Allele by Using RAPD Markers.
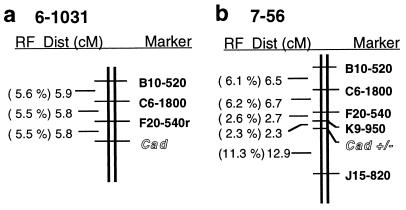
The cad locus and the locus responsible for the low-activity CAD allozyme were mapped independently in two unrelated pine genotypes, using megagametophytes, and both were located in the same genomic region (Fig. 3). We identified RAPD markers linked to the cad locus by bulk segregant analysis in a loblolly pine clone (6-1031) which contained two electrophoretically distinct wild-type CAD allozymes. On the genomic map of clone 7-56 (28), the low CAD activity phenotype associated with the cad-n1 allele mapped to linkage group 5. In clones 6-1031 and 7-56, the cad locus and the locus of the cad-n1 allele each cosegregated with genetic markers common to both individuals. The conserved map order and map distances identified a syntenic region that explains the Mendelian inheritance of these two types of allozyme variation. This result is consistent with genetic control of the low-activity phenotype by the single cad locus of pine (9) and its designation as a mutant cad allele.
Figure 3.
Genetic maps of the cad locus in loblolly pine. Genetic maps of the region flanking the cad locus were generated for clones 6-1031 (a) and 7-56 (b) (on linkage group 5). RF, recombination fraction; Dist (cM), distance in centimorgans; RAPD markers names are Operon primer names followed by the estimated DNA fragment size in base pairs.
Segregation and Phenotype of the cad-n1 Mutant Plants.
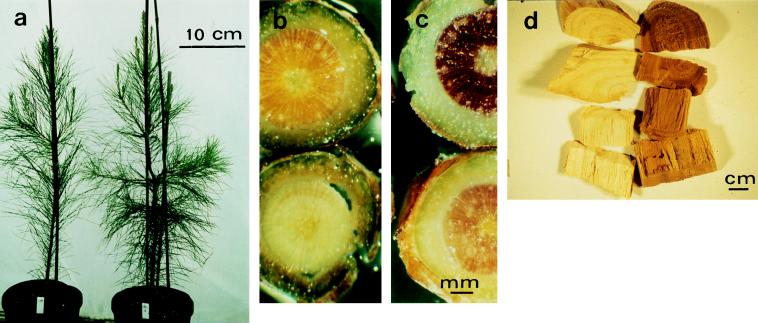
The cad genotype of plants obtained by selfing of clone 7-56 were determined using RAPD markers. cad genotypes could not be determined directly because there was no known DNA sequence polymorphism within the cad gene. Therefore RAPD markers were used to define a multilocus haplotype flanking the cad locus (Fig. 3; ref. 16). The inferred segregation ratio of cad genotypes was 1:2:1, following Mendelian segregation. Under greenhouse conditions, the morphology and growth of cad-n1 homozygotes could not be distinguished easily from that of other genotypes, although several mutants had slightly less straight stems (Fig. 4a). The wood of the cad-n1 homozygotes, however, was distinctly brown, whereas other genotypes had nearly white wood, typical for pine (Fig. 4b).
Figure 4.
Phenotype of the cad-n1 mutant. (a) Seedlings at 10 months, homozygous Cad (left) and cad-n1 (right). (b and c) Stem sections from 7-month-old Cad (lower) and cad-n1 (upper) homozygotes unstained (b) and stained with Weisner reagent (c). (d) Wood chips from 12-year-old trees (homozygous Cad, left; cad-n1, right).
The brown color was evenly distributed throughout the wood of cad-n1 homozygotes and persistent in several 2-year-old saplings (not shown) and even in a 12-year-old tree (Fig. 4d). It was darkest immediately after debarking but was stable in wood samples for several months (Fig. 4d). The color was observed only in tracheids and not in nonlignified ray parenchyma cells, which form lightly colored radial files in xylem cross sections (Fig. 4b). Weisner reagent (10) stained the wood of wild types pink as opposed to dark red for the mutants (Fig. 4c), indicative of an accumulation of cinnamaldehydes (10, 29). Together, the wood color, its cell type distribution, and the intense Weisner stain are consistent with a mutation in lignin biosynthesis that affects CAD activity, resulting in an increase in cinnamaldehyde (14, 30).
Cad Gene Expression.
CAD enzyme activity was assayed in differentiating xylem tissues from 10-month-old pine seedlings by monitoring the oxidation of coniferyl alcohol to coniferaldehyde (11). The wild-type homozygotes had 3.42 ± 0.26 nkat/g of fresh weight tissue, whereas the cad-n1 homozygotes had 0.03 ± 0.05 nkat/g, only 1% on average of wild-type CAD enzyme activity. The reduction of cinnamaldehydes to cinnamyl alcohols is the reaction carried out in vivo by CAD to synthesize monolignols. It was monitored in partially purified xylem protein extracts and, again, cad-n1 homozygotes had between 0% and 1% of the wild-type level of activity. Heterozygosity for the cad-n1 allele resulted in half the wild-type enzyme activity, consistent with heterozygotes having only one active Cad allele.
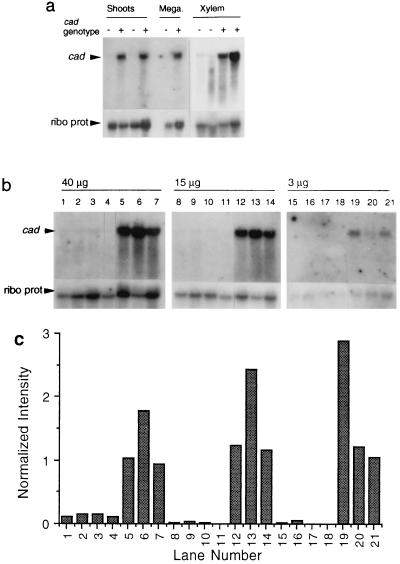
The relative abundance of cad RNA transcript was decreased in tissue samples from mutant plants, including megagametophytes, elongating shoot tips, and differentiating xylem (Fig. 5a). Northern blots were probed with a full-length CAD cDNA probe and a pine ribosomal protein cDNA as an internal standard. The relative abundance of steady-state cad RNA was estimated in total RNA from elongating shoot tips, using 3, 15, and 40 μg from each of several plants (Fig. 5b). It was, on average, 22 times higher in wild-type than in mutant plants, based upon hybridization intensities normalized against the hybridization to the ribosomal probe (Fig. 5c). RNase protection assays with xylem total RNA also showed that the cad message was at least 20 times more abundant in the wild types (not shown). The relative abundance of cad RNA message indicated that the cad-n1 allele is expressed at a very low level, sufficient to account for most of the decrease in CAD enzyme activity.
Figure 5.
Northern analyses of steady-state cad message. (a) RNA blots probed with cad (Upper) and a pine ribosomal protein cDNA (Lower). Samples are total RNA (shoot tips, 10 μg; others, 5 μg) from homozygous or haploid cad-n1 (−) and wild-type (+) genotypes. (b) Total RNAs (40 μg, 15 μg, or 3 μg) from four cad-n1 (lanes 1–4, 8–11, and 15–18) and three wild-type homozygotes (lanes 5–7, 12–14, and 19–21). (c) Histogram of the cad hybridization signal normalized against the ribosomal protein signal for each lane of b.
CAD Substrate Accumulation in cad-n1 Plants.
The total phenolics in ethanol and methanol extracts from finely ground wood were increased 2- to 3-fold in the cad-n1 homozygotes compared with wild-type plants. Extraction with ethanol (Table 1) and successive extractions with methanol (not shown) each gave increases in the relative abundance of phenolics, of similar magnitude, but methanol extracts contained lower concentrations of phenolics. Coniferaldehyde (1b; see Fig. 1), the substrate of CAD, and vanillin were the most abundant extractives, and both increased by at least one order of magnitude in the mutant. Ferulic acid and p-hydroxybenzaldehyde (1a) also increased significantly but were not as abundant. Under alkaline pH, coniferaldehyde reacts to give vanillin and acetaldehyde through a reversed aldol reaction (31). Alkaline hydrolysis of the ethanol extracts increased the ratio of vanillin to coniferaldehyde from 1:1 to 2:1; therefore at least part of the vanillin was derived from accumulated CAD substrate.
Table 1.
Analysis of major ethanol-soluble phenolic extractives
| cad genotype | Total phenolics* | Compound in extract, mg/g dry weight†
|
|||
|---|---|---|---|---|---|
| Coniferaldehyde | Vanillin | p-Hydroxybenzaldehyde | Ferulic acid | ||
| Cad/Cad | 3.3 ± 0.5 | 12.0 | 52.0 | 20.6 | 0.5 |
| Cad/cad-n1 | 2.7 ± 0.7 | ND | ND | ND | ND |
| cad-n1/cad-n1 | 9.1 ± 1.0 | 387.0 | 679.0 | 34.6 | 43.7 |
| Fold increase‡ | 2.8 | 32 | 13 | 1.7 | 87 |
Data are mg of ferulic acid equivalents per g dry weight; mean ± SEM from six plants.
HPLC determination in extracts pooled from four seedlings of each genotype; results are from one typical HPLC analysis. ND, not determined.
Fold increase: cad-n1/cad-n1 value divided by the Cad/Cad value.
Decreased Total Lignin Content in Homozygous cad-n1 Plants.
Total lignin content estimated from Klason and acid-soluble lignin was decreased from 31.7% ± 0.8% of dry weight in the wild type to 28.9% ± 0.5% in the cad-n1 homozygotes (Table 2). In Klason determinations, CWR is treated with sulfuric acid and the residual acid-insoluble material represents most of the lignin (24). However, a small but variable fraction of the lignin is soluble in the acid. In samples from mutant plants, the acid-insoluble lignin was decreased to 91% of wild type, and the acid-soluble lignin remained unchanged.
Table 2.
Effect of the cad-n1 allele on lignin content (% dry weight of CWR)
| cad genotype | Klason method
|
AcBr method*
|
||||
|---|---|---|---|---|---|---|
| Acid-insoluble | Acid-soluble | Total | AcBr | AF | Total | |
| Cad/Cad | 31.0 ± 0.7 | 0.71 ± 0.1 | 31.7 ± 0.8 | 30.7 ± 0.9 | 0.9 ± 0.1 | 31.6 ± 1.0 |
| Cad/cad-n1 | 30.8 ± 0.7 | 0.69 ± 0.1 | 31.5 ± 0.8 | ND | ND | ND |
| cad-n1/cad-n1 | 28.1 ± 0.4 | 0.82 ± 0.1 | 28.9 ± 0.5 | 19.2 ± 0.7 | 8.1 ± 0.2 | 27.3 ± 0.9 |
| Relative content† | 91% | 91% | 86% | |||
CWR were pre-extracted with 1 M NaOH (AF); phenolic content was determined in the AF and lignin content was determined in the AcBr soluble fraction (AcBr). ND, not determined.
Relative content: relative lignin content of cad-n1 homozygotes compared to Cad homozygotes.
We confirmed the small decrease in lignin content by using the AcBr method, in which lignin is determined from the absorbance (A280) of the CWR solubilized in AcBr (26). A saponification pretreatment was needed to completely solubilize the CWR from cad-n1 homozygotes and was used with all genotypes. The phenolics in the alkaline fraction (AF) from the pretreatment of cad-n1 homozygotes accounted for 8% (by weight) of the CWR as opposed to only 0.9% in the wild-type plants (Table 2). The total lignin of the mutant, the sum of the AF and the AcBr-solubilized lignin, was 86% of wild type, consistent with a small decrease in the cad-n1 mutant.
Characterization of the Lignin: Evidence of Changes in Lignin Composition.
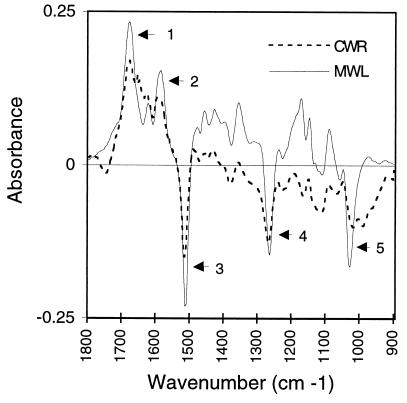
FTIR difference spectra of MWL and of ground wood (CWR) showed an increase in functional groups consistent with an enrichment of coniferaldehyde (1b) subunits in the mutant lignin (Fig. 6, Table 3). MWL is most representative of lignin in plant tissues (22), and FTIR spectra of MWL have been extensively characterized (32). Changes in absorption at several wavelengths in the 1700–1000 cm−1 range provide strong evidence that the MWL from the cad-n1 tree is enriched in carbonyl groups conjugated to aromatic rings, as expected for an increase in coniferaldehyde. The spectra of CWR from mutant plants gave similar results (Fig. 6), consistent with spectra from xylem strips of antisense cad tobacco (33). Conclusions from the MWL sample from the mutant were thus extended to the samples containing the entire cell wall lignin from several plants.
Figure 6.
FTIR difference spectra for MWLs and CWRs. Difference spectra were obtained by subtracting the absorption spectrum of wild type from the spectrum of the mutant: peaks (above the origin) represent stronger absorption in the mutant, troughs are stronger in the wild type. Assignments of absorption bands (10, 32) are 1, 1665–1680 cm−1: carbonyl group (C=O) conjugated to aromatic ring; 2, 1595 cm−1: aromatic ring with C=O stretching; 3, 1510 cm−1: aromatic ring with C—O stretching; 4, 1270 cm−1: C—O stretching, aromatic (methoxyl); 5, 1040 cm−1: dialkyl ether linkages (between cinnamyl alcohol subunits). CWR spectrum is the average from four pairwise comparisons of cad-n1 and wild-type plants.
Table 3.
Diagnostic FTIR absorption ratios of MWL and CWR
| Sample |
A1665/A1510
|
A1595/A1510
|
||
|---|---|---|---|---|
| Cad | cad-n1 | Cad | cad-n1 | |
| MWL | 0.31 | 0.77 | 0.44 | 0.87 |
| CWR | 0.73 | 1.14 | 0.74 | 1.02 |
Materials in this table were Cad and cad-n1 homozygotes.
FTIR absorption ratios A1665/A1510 and A1595/A1510 are diagnostic for cinnamaldehyde content (29, 33) and were indicative of their increase. MWL of the mutant had a 2- to 3-fold increase in these ratios and CWR of mutants had a smaller increase (Table 3). Dehydrogenation lignin polymers (DHPs) can be synthesized in vitro from cinnamyl alcohols (2a, 2b), and addition of cinnamaldehyde (1a, b) to the synthesis can increase the A1665/A1510 and A1595/A1510 ratios of such DHPs (29). A synthesis reaction mixture containing 50% cinnamaldehyde was required to obtain an increase of magnitude similar to that observed with the MWL of cad-n1 plants. Finally, using NaBH4 reduction (34), we estimated that MWL of the cad-n1 mutant contained at least 25% aldehyde as opposed to 10% in the wild type.
DISCUSSION
We have characterized a mutant allele (cad-n1) of the cad gene, discovered in a heterozygous loblolly pine tree through segregation analysis. Genetic mapping of the low-activity CAD allozyme phenotype to the same genomic region as the cad locus is consistent with a single genetic lesion specifically affecting CAD. The phenotype of the vascular tissue is similar to that of bm mutants in maize and related grasses (2, 4, 7, 8). Several bm mutants have been linked to changes in the lignin biosynthetic enzymes COMT (35), CAD (36), or both (14). A mutation in the enzyme ferulate 5-hydroxylase in Arabidopsis thaliana affects lignin composition by reducing the formation of sinapyl alcohol but does not result in a bm phenotype (37).
Recent reports have described the modification of lignin composition using antisense cad genes (5, 14, 30, 33). No decrease in lignin content was detected, even when the residual CAD activity was as low as 7% of wild type (5). The cad-n1 allele has a small effect on lignin content and has lower residual CAD activity than antisense plants and bm mutants described so far. The cad-n1 allele was associated with very low steady-state cad RNA transcript and accumulation of CAD substrate to high levels in xylem. Thus, this allele causes a stronger block in CAD enzyme activity than in previously characterized plants (5, 14, 30).
Reduction or suppression of CAD activity in these diverse plant species has either no detectable effect or a small effect on lignin content, indicating that CAD is normally not rate limiting and has a minor role in regulating metabolic flux through the lignin biosynthetic pathway. However, some minimal level of CAD enzyme is needed to achieve wild-type lignin content, suggesting that a small amount of CAD activity is required for normal flux. In contrast, reduction of phenylalanine ammonia-lyase activity had a more dramatic effect on regulation of flux into this pathway and on lignin content (38).
Mechanisms for the incorporation of cinnamaldehydes directly into the lignin polymer could account for such a small decrease in lignin resulting from decreased CAD activity. Cinnamaldehyde residues normally account for 3–5% of lignin subunits (10, 31), but cell walls of some bm mutants, the pine cad-n1 mutant, and some transgenic antisense cad plants are enriched in such cinnamaldehyde residues (5, 14, 30, 33). The isolated lignin of transgenic poplar (30) and of the pine cad-n1 mutant (ref. 39; this report) provide conclusive evidence of the incorporation of aromatic aldehyde subunits into the lignin polymer at significantly higher levels. Cinnamaldehydes are also efficiently incorporated into in vitro synthesized phenolic dehydrogenation polymers (DHPs), which are dark red (29). In this context, the accumulation of free coniferaldehyde to much higher levels in the cad-n1 mutant is paradoxical; however, incorporation of coniferaldehyde is usually minor in vivo and may become more significant only when its concentration is elevated. The efficiency of incorporation of coniferaldehyde into lignin may be sufficient to maintain flux through the pathway but not enough to reduce the high concentrations of free aldehyde. The extent to which cinnamaldehyde subunits may be increased in lignin and the enzymatic activities involved in their incorporation are unknown.
Alternative routes or mechanisms for the formation of lignin precursors from coniferaldehyde could be utilized or up-regulated in the cad-n1 mutant. It was recently shown by NMR that this mutant incorporates a major proportion of dihydroconiferyl alcohol as a novel lignin precursor (39). Thus, alternative reduction of coniferaldehyde is likely to be carried out by enzymes other than CAD in the formation of modified lignin.
In view of the viability and lignin composition of this mutant, what is the role of CAD? We suggest that CAD may modulate the ratio of cinnamaldehydes to cinnamyl alcohols in lignin. The proportion of aldehyde and alcohol lignin precursors could affect lignin structure (33), properties (30), and ultimately lignin function. Under controlled conditions, cad-n1 mutants did not have aberrant development or growth, suggesting that vascular and mechanical support functions were not significantly impaired. However, it remains to be determined how these functions may be affected under conditions of stress, such as drought. Analysis of the biosynthesis, structure, and functional properties of genetically altered lignin should continue to provide new insights into the structure–function relationship of lignin and its evolution in higher plants.
Acknowledgments
We thank K. Hill, whose work suggested a null CAD allozyme in clone 7-56, and B. Crane, C.-L. Chen, H.-M. Chang, J. Ralph, W. Vermerris, and I. Allona for helpful advice or assistance. The 12-year-old trees were provided by L. Pearson (Westvaco Corporation) and G. Askew (Clemson University). This work was supported by the North Carolina State University Forest Biotechnology Industrial Associates, Grant GN45344–06 from the National Institutes of Health, and Grant DE-FG592ER20085 from the Department of Energy.
ABBREVIATIONS
- AcBr
acetyl bromide
- AF
alkaline fraction
- CAD
cinnamyl alcohol dehydrogenase
- COMT
caffeate/5-hydroxyferulate O-methyltransferase
- CWR
cell wall residue
- FTIR
Fourier-transform infrared
- G6PDH
glucose-6-phosphate dehydrogenase
- MWL
milled wood lignin
- RAPD
random amplified polymorphic DNA
Footnotes
All precusors of lignin are p-hydroxycinnamyl alcohols.
References
- 1.Whetten R W, Sederoff R R. Plant Cell. 1995;7:1001–1013. doi: 10.1105/tpc.7.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boudet A M, Lapierre C, Grima-Pettenati J. New Phytol. 1995;129:203–236. doi: 10.1111/j.1469-8137.1995.tb04292.x. [DOI] [PubMed] [Google Scholar]
- 3.Freudenberg K. Nature (London) 1959;183:1152–1155. doi: 10.1038/1831152a0. [DOI] [PubMed] [Google Scholar]
- 4.Campbell M M, Sederoff R R. Plant Physiol. 1995;110:3–13. doi: 10.1104/pp.110.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halpin C, Knight M E, Foxon G A, Campbell M M, Boudet A M, Boon J J, Chabbert B, Tollier M-T, Schuch W. Plant J. 1994;6:339–350. [Google Scholar]
- 6.Sarkanen K V, Hergert H L. In: Lignins, Occurrence, Formation, Structure and Reactions. Sarkanen K V, Ludwig C H, editors. New York: Wiley Interscience; 1971. pp. 43–94. [Google Scholar]
- 7.Kuc J, Nelson O E. Arch Biochem Biophys. 1964;105:103–113. doi: 10.1016/0003-9861(64)90240-1. [DOI] [PubMed] [Google Scholar]
- 8.Muller L D, Barnes R F, Bauman L F, Colenbrander V F. Crop Sci. 1971;11:413–415. [Google Scholar]
- 9.MacKay J J, Liu W, Whetten R W, Sederoff R R, O’Malley D M. Mol Gen Genet. 1995;247:537–545. doi: 10.1007/BF00290344. [DOI] [PubMed] [Google Scholar]
- 10.Browning B L. Methods of Wood Chemistry. Appleton, WI: Inst. Paper Chem.; 1967. [Google Scholar]
- 11.O’Malley D, Porter S, Sederoff R R. Plant Physiol. 1992;98:1364–1371. doi: 10.1104/pp.98.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabriel O, Gersten D M. Anal Biochem. 1992;293:1–21. doi: 10.1016/0003-2697(92)90036-7. [DOI] [PubMed] [Google Scholar]
- 13.Campbell M M, Ellis B E. Planta. 1992;486:409–417. doi: 10.1007/BF00195322. [DOI] [PubMed] [Google Scholar]
- 14.Pillonel C, Mulder M M, Boon J J, Forster B, Binder A. Planta. 1991;185:538–544. doi: 10.1007/BF00202964. [DOI] [PubMed] [Google Scholar]
- 15.Doyle J J, Doyle J L. Focus. 1987;12:13–15. [Google Scholar]
- 16.Plomion C, Liu B-H, O’Malley D. Theor Appl Genet. 1996;93:1083–1089. doi: 10.1007/BF00230129. [DOI] [PubMed] [Google Scholar]
- 17.Grattapaglia D, Sederoff R. Genetics. 1994;137:1121–1137. doi: 10.1093/genetics/137.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michelmore R W, Paran I, Kesseli R V. Proc Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang S, Puryear J, Cairney J. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- 20.Brown T, MacKey K. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Siedman J G, Smith J A, Struchl K, editors. New York: Wiley; 1997. pp. 4.9.1–4.9.12. [Google Scholar]
- 21.Booker F L, Anttonen S, Heagle A S. New Phytol. 1996;132:483–492. doi: 10.1111/j.1469-8137.1996.tb01868.x. [DOI] [PubMed] [Google Scholar]
- 22.Lundquist K. In: Methods in Lignin Chemistry. Lin S E, Dence C W, editors. Berlin: Springer; 1992. pp. 65–70. [Google Scholar]
- 23.Björkman A. Nature (London) 1954;174:1057–1058. [Google Scholar]
- 24.Effland M J. Tappi. 1977;60:143–144. [Google Scholar]
- 25.Dence C W. In: Methods in Lignin Chemistry. Lin S E, Dence C W, editors. Berlin: Springer; 1992. pp. 33–62. [Google Scholar]
- 26.Iiyama K, Wallis A F A. Wood Sci Technol. 1988;22:271–280. [Google Scholar]
- 27.Gifford M, Foster A S. Morphology and Evolution of Vascular Plants. 3rd Ed. New York: Freeman; 1989. [Google Scholar]
- 28.O’Malley D M, Grattapaglia D, Chaparro J X, Wilcox P L, Amerson H V, Liu B-H, Whetten R, McKeand S E, Kuhlman E G, McCord S, Crane B, Sederoff R R. In: Genomes of Plants and Animals: 21st Stadler Genetics Symposium. Gustafson J P, Flavell R B, editors. New York: Plenum; 1995. pp. 87–102. [Google Scholar]
- 29.Higuchi T, Ito T, Umezawa T, Hibino T, Shibata D. J Biotechnol. 1994;37:151–158. [Google Scholar]
- 30.Baucher M, Chabbert B, Pilate G, van Doorsselaere J, Tollier M-T, Petit-Conil M, Cornu D, Monties B, van Montagu M, Inzé D, Jouanin L, Boerjan W. Plant Physiol. 1996;112:1479–1490. doi: 10.1104/pp.112.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hrutfiord B F. In: Lignins, Occurrence, Formation, Structure and Reactions. Sarkanen K V, Ludwig C H, editors. New York: Wiley Interscience; 1971. pp. 487–509. [Google Scholar]
- 32.Faix O. In: Methods in Lignin Chemistry. Lin S E, Dence C W, editors. Berlin: Springer; 1992. pp. 81–109. [Google Scholar]
- 33.Stewart D, Yahiaoui N, McDougall G J, Myton K, Marque C, Boudet A M, Haigh J. Planta. 1997;201:311–318. doi: 10.1007/s004250050072. [DOI] [PubMed] [Google Scholar]
- 34.Chen C-L. In: Methods in Lignin Chemistry. Lin S E, Dence C W, editors. Berlin: Springer; 1992. pp. 446–457. [Google Scholar]
- 35.Vignols F, Rigau J, Torres M A, Capellades M, Puigdomènech P. Plant Cell. 1995;7:407–416. doi: 10.1105/tpc.7.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halpin C, Foxon G A, Fentem P A. In: Biotechnology in Animal Feeds and Animal Feeding. Wallace R J, Chesson A, editors. New York: VCH; 1995. pp. 279–293. [Google Scholar]
- 37.Meyer K, Cusumano J C, Somerville C, Chapple C C S. Proc Natl Acad Sci USA. 1996;93:6869–6874. doi: 10.1073/pnas.93.14.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bate N J, Orr J, Ni W, Meromi A, Nadler-Hassar T, Doerner P W, Dixon R A, Lamb C J, Elkind Y. Proc Natl Acad Sci USA. 1994;91:7608–7612. doi: 10.1073/pnas.91.16.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ralph, J., MacKay, J. J., Hatfield, R. D., O’Malley, D. M., Whetten, R. W. & Sederoff, R. R. (1997) Science, in press. [DOI] [PubMed]