Abstract
Bacterial adhesion and the subsequent formation of biofilm are major concerns in biotechnology and medicine. The initial step in bacterial adhesion is the interaction of cells with a surface, a process governed by long-range forces, primarily van der Waals and electrostatic interactions. The precise manner in which the force of interaction is affected by cell surface components and by the physiochemical properties of materials is not well understood. Here, we show that atomic force microscopy can be used to analyze the initial events in bacterial adhesion with unprecedented resolution. Interactions between the cantilever tip and confluent monolayers of isogenic strains of Escherichia coli mutants exhibiting subtle differences in cell surface composition were measured. It was shown that the adhesion force is affected by the length of core lipopolysaccharide molecules on the E. coli cell surface and by the production of the capsular polysaccharide, colanic acid. Furthermore, by modifying the atomic force microscope tip we developed a method for determining whether bacteria are attracted or repelled by virtually any biomaterial of interest. This information will be critical for the design of materials that are resistant to bacterial adhesion.
Bacterial adhesion onto inanimate surfaces is a critical issue in processes ranging from the biofouling of industrial equipment to dental decay to infections of biomaterials for medical use. Bacterial infections associated with the formation of biofilms refractile to antibiotic therapy is one of the main reasons for the failure of devices such as catheters, vascular grafts, joint prostheses, and heart valves (1–3). The first step in bacterial adhesion is the immediate attachment of bacteria onto a substratum which is a reversible, nonspecific process (3–5). This initial interaction between bacteria and artificial surfaces is a key determinant in biofilm formation. If the approach of bacteria to a surface is unfavorable, cells must overcome an energy barrier to establish direct contact with the surface. Only when bacteria are in close proximity to the surface do short-range interactions become significant. Thereafter, protein-ligand–binding events mediated by a plethora of microbial adhesins and in some cases the production of extracellular polymers render the binding process practically irreversible (6).
Initial bacterial attraction or repulsion to a particular surface can be described in terms of colloidal interactions. Consequently, the force of interaction depends on physiochemical parameters such as surface-free energy and charge density (7–11). The propensity of bacteria to adhere onto surfaces has been estimated by counting the number of bacteria that remain attached to surfaces following incubation for a specified length of time (5, 12). This approach is qualitative, time consuming, and has low sensitivity. Moreover, the resulting number of adherent bacteria is determined by multiple factors in addition to long-range attractive/repulsive interactions. A direct and quantitative means for specifically measuring the long-range interactions between bacteria and surfaces can provide important information to direct the design of materials refractile to bacterial adhesion and for the control of biofilm formation.
Atomic force microscopy (AFM) has been used extensively to probe the interactions of colloidal particles with planar surfaces (13–17). With respect to biological applications, AFM has been used to detect forces in the piconewton range while operating under physiological conditions. Biological interactions that have been investigated via AFM include antibody-antigen recognition, protein-ligand binding, and complementary DNA base pairing (18–22). In this work, we show that AFM is an exquisitely sensitive tool for analyzing whether long-range interactions between bacteria and surfaces are attractive or repulsive and for understanding the nature of the underlying forces. Moreover, by combining the sensitivity of AFM with the use of well-characterized isogenic strains differing in cell surface composition, we have succeeded in dissecting the contribution of specific cell surface components to the forces of interaction.
MATERIALS AND METHODS
Strains and Growth Conditions.
Escherichia coli K-12 mutants deficient in lipopolysaccharide (LPS) synthesis were obtained from the E. coli Genetic Stock Center (Department of Biology, Yale University, New Haven, CT). The parental strain D21 synthesizes intact core LPS molecules whereas the isogenic mutants D21e7, D21f1, and D21f2 contain progressively truncated LPS (23, 24). D21e19 lacks a branched galactose residue (25). SG22147 (MC4100 lon+) and the mutant SG21011 (SG22147 rcsC137 ompC∷Tn5) were kindly provided by Dr. Susan Gottesman (National Cancer Institute, Bethesda, MD). Mutations that inactivate rcsC, a negative regulator of capsule synthesis, result in the overproduction and secretion of colanic acid (26, 27). All strains were grown aerobically in Luria broth at 37°C.
Cell Immobilization.
Polyethyleneimmine- (PEI; Mr 1200) coated glass was prepared by soaking 1- × 1-cm glass slips in 1 M HNO3 overnight, rinsing with water followed by methanol, and air dried. A drop of 1% PEI was placed on one side of the glass and allowed to adsorb for 3 hr, after which the drop was decanted. Glass slides were rinsed in water and stored at 4°C. Similarly, a drop of PEI solution was placed on silicon nitride tips and allowed to adsorb for 2.5 hr. Excess solution was decanted and cantilevers were rinsed in water and stored at 4°C.
Cells were harvested in mid-exponential phase by centrifugation at 8,000 rpm for 10 min. Cells were washed in PBS (pH 7.2) and then stirred in 2.5% vol/vol glutaraldehyde for 2 hr at 4°C at a final concentration of 0.6–0.8 mg dry cell weight/ml (28). Control experiments consisted of incubating cells with either 5% vol/vol glutaraldehyde for 2 hr or 1.5% vol/vol glutaraldehyde for 4 hr. Prior to use, glutaraldehyde was purified by stirring a 2.5 or 10% vol/vol solution with 50 mg/ml charcoal at 4°C for 24 hr (28). After fixing with glutaraldehyde, the cells were rinsed and resuspended in 1 mM Tris (pH 7.5). The cell suspension was incubated at 4°C overnight. Cells were rinsed repeatedly and resuspended in either in 1 mM Tris or distilled deionized water (pH 6.5).
To immobilize the bacteria onto a rigid substrate, a drop of the glutaraldehyde-treated cell suspension was placed on PEI-coated glass which was placed in a vacuum desiccator at room temperature to evaporate excess water (2–3 hr); the cells themselves were not desiccated. To immobilize bacteria onto cantilevers, a pellet of cells was manually transferred onto PEI-coated tips. The pellet was further treated with an additional drop to 2.5% vol/vol glutaraldehyde and samples were incubated at 4°C for 1–2 hr. Cantilevers were then rinsed in water and air dried. Scanning electron microscopy was performed on all tips coated with bacteria after AFM measurements to verify the presence of bacteria on the silicon nitride tip.
AFM Operation.
A Nanoscope III Contact Mode AFM (Digital Instruments, Santa Barbara, CA) was used to image cells and measure forces of interaction. Nanoprobe silicon nitride cantilevers with a spring constant of k = 0.06 N/m were obtained from Digital Instruments. New, clean cantilevers were used for every force measurement. Experiments were conducted in a fluid cell filled with distilled deionized water (pH 6.5), 1 mM Tris (pH 7.5), or 1 mM Tris plus 100 mM NaCl (pH 7.5). Force measurements were carried out by engaging the AFM without touching the surface to prevent any tip contamination from the sample. The tip was then approached to the surface in 100-nm increments with the specified Z scan size of 300 nm at a frequency of 1 Hz. Surfaces were imaged after every force curve to confirm the presence of a continuous bacterial lawn. Silicon nitride tips were checked for cracks or breaks under an optical microscope before and after force measurements.
Force curves recorded as the tip approached the surface were analyzed to determine the initial interactions between surfaces and bacteria. All force curves were normalized so that the tip deflection was 0 nm where there was no interaction, and the slope of the constant compliance region (portion of curve where cantilever moves with the surface) was equal to the rate of piezo displacement. Representative force curves for all bacterial strains were plotted together by aligning the zero deflection and constant compliance portions of the curves.
Zeta Potential and Contact Angle Measurements.
Zeta potential was determined for cells with and without glutaraldehyde treatment. Cells were harvested in mid-exponential phase, washed, and resuspended in 1 mM Tris (pH 7.5). Zeta potential was measured using a Zeta Reader (model ZR12s, Komline Sanderson, Peapack NJ) with the electric field set to 10, 20, 30, and 40 V/cm.
Contact angles were measured with both stationary and exponential phase cultures by first filtering cells resuspended in either 1 mM Tris (pH 7.5) or 1 M PBS (pH 7.2) onto 0.45-μm pore diameter Type HV Millipore membranes under vacuum for 20–40 min. Contact angles of the resulting cell layer on the membrane filter surface were measured with a goniometer (Rame-Hart, Inc., Mountain Lakes, NJ) using distilled deionized water and diiodomethane (99%, Mr 267.84).
Polymethylmethacrylate (PMMA) Substrate Preparation.
PMMA (Mr 330,000) was dissolved in toluene to a final concentration of 2% wt/vol. Glass was first cleaned with acetone. A thin film of hexanemethyldisilane (97% vol/vol) was spun on the glass followed by a thin polymer film of PMMA at 2,000 rpm. Samples were dried on a hot plate at 90°C for 10 min.
RESULTS
Sample Preparation for AFM Measurements.
To perform AFM measurements, it was first necessary to establish a reliable and mild procedure for the formation of confluent bacterial lawns on the surface of a suitable support. In the absence of a confluent cell layer, the AFM would have to be used in imaging mode prior to force measurements to locate the bacteria. However, tip contamination during imaging could affect the subsequent force measurements, resulting in artifacts. The formation of a confluent bacterial lawn on the support surface ensures that the AFM tip interacts with the bacteria and circumvents the need for prior imaging of the cells.
Lab strains of E. coli do not adhere well onto glass. A number of coatings used to promote cell adhesion including poly-l-lysine, silane, chitosan, and Cell-Tak were evaluated, but none was capable of generating a confluent bacterial layer on the support. The optimal approach for producing uniform bacterial lawns suitable for AFM measurements was to harvest cells in mid-exponential phase, fix them in 2.5% vol/vol glutaraldehyde, and finally adsorb cells onto PEI-coated glass. Treatment with glutaraldehyde was found to be essential for the covalent immobilization of E. coli onto PEI-coated glass surfaces. In this study, we investigated the contribution of LPS and colanic acid to bacterial adhesion. Both of these macromolecules lack free amines and hence are not chemically modified in any way by glutaraldehyde. Moreover, the following evidence argues that glutaraldehyde does not affect the overall physiochemical properties of the bacterial cell surface responsible for long-range interactions: (i) Contact angle measurements which reflect the surface free energy of the bacteria (7, 11) revealed no differences between cells before and after glutaraldehyde treatment. For example, contact angles measured with water as a polar fluid and diiodomethane as a nonpolar fluid (7) on a lawn of E. coli D21 deposited on 0.45-μm pore size filters were 27 ± 7° and 43 ± 3°, respectively. Following glutaraldehyde treatment, the corresponding contact angles were 26 ± 3° and 49 ± 4°, respectively. (ii) Zeta potentials measured for wild-type D21 cells before and after glutaraldehyde treatment were −28.8 ± 0.7 mV and −28.9 ± 1.7 mV, respectively. Thus, the surface charge density of the cells was not affected by glutaraldehyde treatment. (iii) Finally, changing glutaraldehyde concentration did not have any effect on AFM force measurements. Force curves for D21 and D21f2 fixed with either 2.5% vol/vol glutaraldehyde for 2 hr, 5% vol/vol glutaraldehyde for 2 hr, or 1.5% vol/vol glutaraldehyde for 4 hr were indistinguishable (data not shown).
Fig. 1 shows a representative image of a lawn of E. coli D21e7. The lawn is confluent, ensuring that as the cantilever approaches the surface it interacts with the bacteria and not with the underlying glass. Individual bacteria are readily distinguishable. Not surprisingly, atomic resolution of E. coli surface molecules and structures was not possible due to the fluidity of the membrane (29, 30).
Figure 1.
AFM Image of E. coli D21e7 lawn immobilized on glass surface. Cells were harvested in mid-exponential phase, fixed with glutaraldehyde, and adsorbed onto PEI-coated glass. A 10- × 10-μm scan was performed in 1 mM Tris using the long thin cantilever at a scan rate of 15.26 Hz.
Force Measurements.
AFM force measurements were first used to evaluate the significance of LPS composition to the interaction of E. coli with the silicon nitride tip. The outer membrane of E. coli and other Gram-negative bacteria is an asymmetric lipid bilayer whose inner layer is composed of phospholipids, whereas the only lipid in the external layer is LPS (31, 32). LPS is an important virulence factor as well as a significant determinant in the adhesion of bacteria to nonbiological surfaces and tissues (33–36). LPS of E. coli consists of three regions: lipid A, core polysaccharide, and O antigen (37, 38). Lipid A, which anchors LPS molecules to the outer leaflet of the outer membrane, is covalently attached to the core polysaccharide molecule (36). The core polysaccharide is an oligosaccharide chain 20 Å in length (Fig. 2A) (37). The O antigen, which is not synthesized in E. coli K-12 strains, consists of repeating branched polymers of sugar residues attached to the core polysaccharide molecule (37, 39).
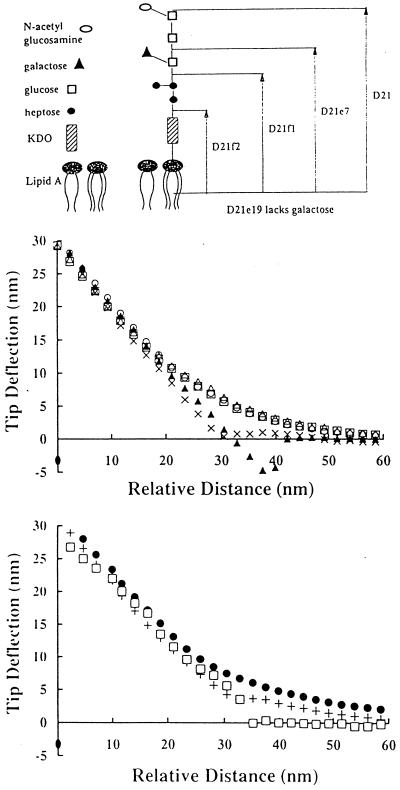
Figure 2.
Interaction between AFM tip and lawns of isogenic E. coli strains differing in LPS composition. (A) Core LPS structure of E. coli K12 strain D21 and its isogenic LPS mutants expressing truncated core LPS molecules. (B) Representative force curves between bare silicon nitride tip and isogenic E. coli mutant strains expressing truncated core LPS molecules in 1 mM Tris (pH 7.5). ▴, D21; ×, D21e7; □, D21e19; ○, D21f1; ▵, D21f2. (C) Representative force curves between bare silicon nitride tip and D21f2 in distilled deionized water, 1 mM Tris, and 1 mM Tris plus 100 mM NaCl. •, D21f2 in water + D21f2 in Tris; □, D21f2 in Tris + NaCl.
In this work, we used a set of isogenic E. coli K-12 strains carrying mutations in the genes involved in LPS synthesis. These mutations result in LPS molecules with progressively truncated core polysaccharide structures (Fig. 2A). D21e19 reportedly lacks only a branched galactose residue (25). Although the LPS structure of D21e19 is not known with certainty, it has been included in this study for the sake of completion. Outer membrane fractions from the isogenic E. coli strains were isolated by differential detergent extraction (40, 41) and proteins were resolved electrophoretically as described by Lugtenberg et al. (42). No differences in outer membrane protein composition or relative concentrations were detected for strains D21, D21e7, D21f1, and D21f2 (A.R. and G.G., unpublished data). In contrast, a prominent band (Mr 68,000) not present in any of the other strains was evident in D21e19. Thus, with the exception of D21e19, the only detectable difference in the surface topography of the four mutants appears to be in the structure of LPS.
AFM measurements were performed in either distilled deionized water, 1 mM Tris buffer, or 1 mM Tris plus 100 mM NaCl. Clean tips were used for every force measurement. The AFM was engaged without touching the surface so as to avoid tip contamination, and then lowered toward the surface in increments of 100 nm with a scan size of 300 nm/s. At least four force curves were reproducibly obtained for each cell sample. Control experiments were performed to rule out the possibility that the PEI coating the glass substrate can adsorb onto the silicon nitride tip and affect force measurements.
Typical tip displacement versus approach distance curves for the five isogenic LPS mutant strains in 1 mM Tris are shown in Fig. 2B. As the tip approached the bacterial lawns of D21e19, D21f1, and D21f2, it experienced a repulsive force with no attractive component. This is not unexpected since both the silicone nitride tip (16, 43) and the E. coli cell surface are negatively charged in aqueous solution. E. coli is negatively charged in part due to the ketodeoxyoctonoic acid moiety present at the base of the LPS molecule. The force curves for D21 and D21e7 were statistically different from those obtained for D21e19, D21f1, and D21f2. Strains D21 and D21e7 did not repel the silicon nitride tip, presumably because the longer LPS chains shield the negatively charged residues proximal to the cell surface, thus dampening electrostatic repulsion. In fact, D21 exhibited a mild attraction for the cantilever tip.
Force measurements for D21f2, the mutant with the most severely truncated LPS, in water, 1 mM Tris, and 1 mM Tris plus 100 mM NaCl are shown in Fig. 2C. The repulsion of D21f2 decreased in the presence of Tris buffer and was completely eliminated by the addition of NaCl. Force curves obtained in the presence of NaCl were not smooth, consistent with observations by Senden and Drummond (43). Conversely, the cantilever tip experienced a net attraction to D21 cells regardless of the presence of buffer or salt (data not shown). These results reveal that the repulsion between the cantilever tip and E. coli mutants having truncated LPS is governed by electrostatic interactions. Consistent with this model, zeta potential measurements revealed that D21f2 is more electronegative relative to wild-type D21 strain (−42.3 ± 4.0 mV and −28.9 ± 1.7 mV, respectively).
Capsular polysaccharides are secreted by both Gram-negative and Gram-positive bacteria to form a highly hydrated, negatively charged, protective glycocalyx surrounding the bacterium (44). Unlike extracellular polysaccharides such as alginate, involved in bacterial adhesion and biofilm formation of Pseudomonas aeruginosa (45, 46), capsular polysaccharides help prevent immunological recognition, phagocytosis, and adhesion (5). Under conditions of stress, E. coli synthesizes colanic acid, a repeating polymer of glucose, galactose, fucose, glucuronic acid, and pyruvate forming a capsule around the E. coli cell surface (26, 31). The effect of colanic acid synthesis on adhesion was examined by comparing the wild-type strain SG22147 with SG21011, an isogenic mutant deficient in the rscC gene which encodes a negative regulator of colanic acid synthesis (27). SG21011 constitutively overproduces colanic acid, resulting in a mucoid morphology. Force measurements were performed with the parent and colanic acid mutant strains in both distilled deionized water and 1 mM Tris. The interactions of both strains with the AFM tip were repulsive (A.R., Y.L.O., M.M.S., and G.G., submitted for publication). No difference in the magnitude of the repulsive force exerted by the parental and mutant strains could be distinguished in distilled deionized water. However, in the presence of buffer, the mutant overproducing colanic acid repelled the AFM tip to a greater extent than the parent strain, consistent with the higher negative charge density of the capsular material (data not shown).
Force Measurements Between Bacteria and Biomaterials.
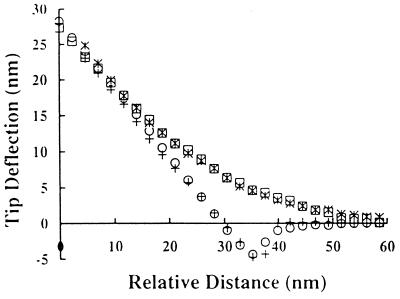
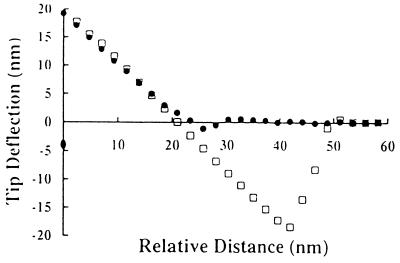
A number of groups have used AFM to analyze the interaction between planar surfaces and materials other than silicon nitrite tips (13–16). For this purpose, microspheres and even metallic shards can be directly attached onto AFM tips and used to probe planar surfaces. Similarly, we attached glass and polystyrene microspheres onto cantilevers and used them to probe bacterial lawns (A.R., Y.L.O., M.M.S., and G.G., submitted for publication). However, this approach is limited by the lack of relevant materials that can be manufactured as microspheres 10–30 μm in diameter. Alternatively, we coated cantilever tips with a confluent layer of bacteria (Fig. 3). Modified cantilevers were used to obtain force curves with planar glass surfaces. Due to similarities in surface characteristics for glass and silicon nitrite (43), the force of interaction between a silicon nitride tip and a lawn of bacteria was expected to be similar to the interaction between an AFM tip coated with bacteria and a clean glass surface. Indeed, the force curves for each of the described configurations were experimentally indistinguishable for D21 and D21f2 (Fig. 4). These results demonstrate that when the AFM tip is uniformly coated with bacteria, the resulting force measurements do indeed reflect the interaction between bacteria and the substrate being probed. In this mode AFM can be readily employed to measure the interaction of bacteria with different materials of biomedical and biotechnological interest. For example, Fig. 5 shows force curves for a PMMA substrate probed by AFM tips coated with the wild-type E. coli strain D21 and the truncated LPS mutant strain D21f2. PMMA is used extensively in blood pumps, dialysis systems, contact lenses, and dentures (47). No electrostatic repulsion was detected between D21f2 and PMMA. On the other hand, D21 was strongly attracted to PMMA. This attractive force was not affected by the addition of salt (Y.L.O., A.R., M.M.S., and G.G., manuscript in preparation).
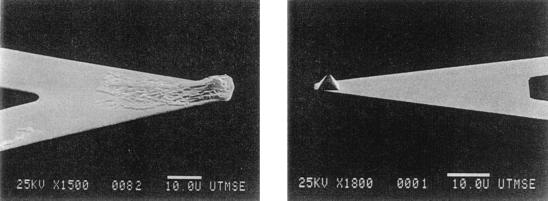
Figure 3.
Scanning electron microscopic images of bare silicon nitride tip and silicon nitride tip coated with E. coli D21. Bar, 10 μm.
Figure 4.
Comparison of force curves obtained by approaching bare silicon nitride tips to bacterial lawns immobilized on glass with force curves obtained by approaching bacterial-coated tips to clean glass surfaces. All measurements were performed in 1 mM Tris (pH 7.5). +, bare silicon nitride tip probing D21 bacterial lawn; ○, silicon nitride tip coated with D21 probing glass substrate; ≍⃒, bare silicon nitride tip probing D21f2 bacterial lawn; □, silicon nitride tip coated with D21f2 probing glass substrate.
Figure 5.
Average force curves (n = 4) between silcon nitride tips coated with E. coli D21 and D21f2 and glass spin-coated with PMMA. Measurements were performed in 1 mM Tris (pH 7.5). □, D21; •, D21f2.
DISCUSSION
In this report, we show that the atomic force microscope represents a highly sensitive and versatile tool for determining the interactions between bacteria and surfaces. Force measurements with Gram-negative bacteria were possible because the rigid peptidoglycan layer present in the cell wall allows the cells to resist elastic deformation by the AFM tip (48, 49). The absence of elastic deformation is evident by the fact that the constant compliance region of the force curves (the region where the sample and cantilever are in contact and move together) is identical to that expected for a rigid sample (data not shown; refs. 50 and 51). In contrast, mammalian cell membranes are readily deformable (49, 50, 52).
The AFM-based methodology was shown to be sensitive enough to dissect the effect of subtle changes in overall bacterial cell surface composition on the initial interaction of bacteria with various materials. Specifically, the sensitivity of force measurements allowed the comparison of well-characterized isogenic mutant strains differing only in the composition of specific cell surface components. As shown in Fig. 2B, it was possible to discern differences in the force of interaction among isogenic strains differing only in the composition and length of core LPS. Unlike their isogenic wild-type counterparts, cells with truncated LPS molecules were not attracted to the AFM tip. In fact, progressively shorter LPS molecules resulted in increasing electrostatic repulsion to the negatively charged silicon nitride tip. Consistent with this data, the zeta potential of D21f2, the mutant expressing the shortest LPS molecule, was substantially more electronegative than that of the wild-type strain. As expected, the magnitude of this electrostatic repulsion was reduced in the presence of buffer and salt (Fig. 2C). A similar behavior was observed with other negatively charged materials such as mica (Y. L.O., A.R., G.G., and M.M.S., manuscript in preparation). The most plausible explanation for these results is that in wild-type cells the longer LPS molecules serve to shield the negatively charged ketodeoxyoctonoic moiety. As a result, attractive van der Waals interactions dominate and the bacteria experience a net attraction to the surface.
Electrostatic interactions were also found to dominate the adhesive behavior of bacteria that overproduce colanic acid. In water, the force curves for the parent strain and for the colanic acid-producing mutant strain were indistinguishable, whereas in buffer, the mutant strain exhibited a significantly greater repulsion to the silicon nitride tip than the parental strain. In this case the parental strain SG22147 did not exhibit an attractive interaction with the silicon nitride tip in contrast to the wild-type D21 strain. This was not unexpected since SG22147 has a different lineage and hence a different cell envelope and LPS composition than D21.
By forming a confluent bacterial layer directly onto the cantilever tip, it was possible to use AFM to determine interactions between bacteria and biomaterials such as PMMA. Modifying the cantilever tip with a confluent layer of bacteria is straightforward and allows AFM to be readily employed as a means of screening biomaterials in terms of their susceptibility to bacterial adhesion and infections. For example, this technique has allowed, for the first time, the determination of whether commonly used materials (e.g., polystyrene, Teflon) repel or attract bacteria based on different bacterial cell and biomaterial surface properties. The quantitative nature of force versus distance curves obtained in AFM measurements has been found to be very useful in not only the qualitative understanding of bacterial adhesion, but also in the quantitative evaluation of different mathematical models that account for electrostatic, dispersion, hydrophobic, and steric interactions (Y.L.O., A.R., M.M.S., and G.G., manuscript in preparation).
Acknowledgments
We are grateful to Professor A. Freemam (Tel-Aviv University, Tel-Aviv, Israel) for advice regarding E. coli immobilization, Dr. Susan Gottesman for providing E. coli strains, and Daren L. Stephens for help with OM protein isolation. We would also like to thank Professors W. Pitt and R. McLean for helpful comments. Anneta Razatos is a Whitaker Foundation Graduate Fellow. This work was supported by Grant R821 268-01-0 from the Environmental Protection Agency (to G.G. and M.M.S.).
ABBREVIATIONS
- AFM
atomic force microscopy
- LPS
lipopolysaccharide
- PEI
polyethyleneimmine
- PMMA
polymethylmethacrylate
References
- 1.Stickler D J, McLean R J C. Cells Mater. 1995;5:167–182. [Google Scholar]
- 2.Bryers J D. Biotechnol Progr. 1987;3:57–68. [Google Scholar]
- 3.Gristina A G. Science. 1987;237:1588–1595. doi: 10.1126/science.3629258. [DOI] [PubMed] [Google Scholar]
- 4.Christensen G D, Baldassarri L, Simpson W A. Methods Ezymol. 1995;253:477–500. doi: 10.1016/s0076-6879(95)53040-1. [DOI] [PubMed] [Google Scholar]
- 5.Ofek I, Doyle R J. Bacterial Adhesion to Cells and Tissues. New York: Chapman & Hall; 1994. [Google Scholar]
- 6.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 7.Oss C J v. Biofouling. 1991;4:25–35. [Google Scholar]
- 8.Klotz S A. In: Microbial Cell Surface Hydrophobicity. Doyle R J, Rosenberg M, editors. Washington, DC: Am. Soc. Microbiol.; 1990. pp. 107–136. [Google Scholar]
- 9.Loosdrecht M C M v, Lyklema J, Norde W, Schraa G, Zehnder A J B. Appl Environ Microbiol. 1987;53:1898–1901. doi: 10.1128/aem.53.8.1898-1901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mozes N, Marchal F, Hermesse M P, Haecht J L V, Reuliaux L, Leonard A J, Rouxhet P G. Biotechnol Bioeng. 1987;30:439–450. doi: 10.1002/bit.260300315. [DOI] [PubMed] [Google Scholar]
- 11.Busscher H J, Weerkamp A H, Mei H C v d, Pelt A W J v, Jong H P D, Arends J. Appl Environ Microbiol. 1984;48:980–983. doi: 10.1128/aem.48.5.980-983.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher M, Pringle J H. J Colloid Interface Sci. 1985;140:5–13. [Google Scholar]
- 13.Yoon R-H, Flinn D H, Rabinovich Y I. J Colloid Interface Sci. 1997;185:363–370. doi: 10.1006/jcis.1996.4583. [DOI] [PubMed] [Google Scholar]
- 14.Basu S, Sharma M M. J Colloid Interface Sci. 1996;181:443–455. [Google Scholar]
- 15.Biggs S. Langmuir. 1995;11:156–162. [Google Scholar]
- 16.Butt H-J, Jaschke M, Ducker W. Bioelectrochem Bioenerg. 1995;38:191–201. [Google Scholar]
- 17.Ducker W A, Xu Z, Israelachvili J N. Langmuir. 1994;10:3279–3289. [Google Scholar]
- 18.Browning-Kelly M E, Wadu-Mesthrige K, Hari V, Liu G Y. Langmuir. 1997;13:343–350. [Google Scholar]
- 19.Boland T, Ratner B D. Proc Natl Acad Sci USA. 1995;92:5297–5301. doi: 10.1073/pnas.92.12.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chilkoti A, Boland T, Ratner B D, Stayton P S. Biophys J. 1995;69:2125–2130. doi: 10.1016/S0006-3495(95)80083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Florin E-L, Moy V T, Gaub H E. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 22.Lee G U, Chrisey L A, Colton R J. Science. 1994;266:771–773. doi: 10.1126/science.7973628. [DOI] [PubMed] [Google Scholar]
- 23.Havekes L, Tommassen J, Hoekstra W, Lugtenberg B. J Bacteriol. 1977;129:1–8. doi: 10.1128/jb.129.1.1-8.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boman H G, Monner D A. J Bacteriol. 1975;121:455–464. doi: 10.1128/jb.121.2.455-464.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eriksson-Grennberg K G, Nordström K, Englund P. J Bacteriol. 1971;108:1210–1223. doi: 10.1128/jb.108.3.1210-1223.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottesman S, Stout V. Mol Microbiol. 1991;5:1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x. [DOI] [PubMed] [Google Scholar]
- 27.Stout V, Gottesman S. J Bacteriol. 1990;172:659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freeman A, Abramov S, Georgiou G. Biotechnol Bioeng. 1996;52:625–630. doi: 10.1002/(SICI)1097-0290(19961205)52:5<625::AID-BIT10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 29.Hansma H G, Hoh J H. Annu Rev Biophys Biomol Struct. 1994;25:115–139. doi: 10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- 30.Mulhern P J, Blackford B L, Jericho M H, Southam G, Bereridge T J. Ultramicroscopy. 1992;42–44:1214–1221. [Google Scholar]
- 31.Nikaido H, Vaara M. In: Escherichia coli and Salmonella typherium: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol. Press; 1987. pp. 7–22. [Google Scholar]
- 32.Lugtenberg B, Alphen L v. Biochim Biophys Acta. 1983;737:51–115. doi: 10.1016/0304-4157(83)90014-x. [DOI] [PubMed] [Google Scholar]
- 33.Klein N J, Ison C A, Peakman M, Levin M, Hammerschmidt S, Frosch M, Heyderman R S. J Infect Dis. 1996;173:172–179. doi: 10.1093/infdis/173.1.172. [DOI] [PubMed] [Google Scholar]
- 34.Elgavish A. Infect Immun. 1993;61:3304–3312. doi: 10.1128/iai.61.8.3304-3312.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valkonen K H, Veijola J, Dagberg B, Uhlin B E. Mol Microbiol. 1991;5:2133–2141. doi: 10.1111/j.1365-2958.1991.tb02143.x. [DOI] [PubMed] [Google Scholar]
- 36.Raetz C R H. In: Escherichia coli and Salmonella typherium: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: ASM Press; 1987. pp. 498–503. [Google Scholar]
- 37.Escherichia coli and Salmonella typherium: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtis, R., III, Ingraham, J. L., Lin, E. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (1996) (ASM Press, Washington, DC).
- 38.Rooney S A, Goldfine H. J Bacteriol. 1972;111:531–541. doi: 10.1128/jb.111.2.531-541.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rick P D. In: Escherichia coli and Salmonella typherium: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: ASM Press; 1987. pp. 648–662. [Google Scholar]
- 40.Filip C, Fletcher G, Wulff J L, Earhart C F. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inouye M, Guthrie J P. Proc Natl Acad Sci USA. 1969;64:957–961. doi: 10.1073/pnas.64.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lugtenberg B J, Meyers J, Peters R, Hoek P v d, Alphen L v. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 43.Senden T J, Drummond C R. Colloids Surf. 1995;94:29–51. [Google Scholar]
- 44.Roberts I S. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 45.Schurr M J, Yu H, Boucher J C, Hibler N S, Deretic V. J Bacteriol. 1995;177:5670–5679. doi: 10.1128/jb.177.19.5670-5679.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deretic V, Marin M W, Schurr M J, Mudd M H, Hibler N S, Curcic R, Boucher J C. Bio/Technology. 1993;11:1133–1136. doi: 10.1038/nbt1093-1133. [DOI] [PubMed] [Google Scholar]
- 47.Lee H B, Kim S S, Khang G. In: The Biomedical Engineering Handbook. Bronzino J D, editor. Boca Raton, FL: CRC; 1995. pp. 581–597. [Google Scholar]
- 48.Brock T D, Madigan M T, Martinko J M, Parker J. Biology of Microorganisms. Englewood Cliffs, NJ: Prentice–Hall; 1994. [Google Scholar]
- 49.Hoh J H, Schoenenberger C-A. J Cell Sci. 1994;107:1105–1114. doi: 10.1242/jcs.107.5.1105. [DOI] [PubMed] [Google Scholar]
- 50.Goldmann W H, Ezzell R M. Exp Cell Res. 1996;226:226–237. doi: 10.1006/excr.1996.0223. [DOI] [PubMed] [Google Scholar]
- 51.Radmacher M, Fritz M, Cleveland J P, Walters D A, Hansma P K. Langmuir. 1994;10:3809–3814. [Google Scholar]
- 52.Schoenenberger C A, Hoh J H. Biophys J. 1994;67:929–936. doi: 10.1016/S0006-3495(94)80556-9. [DOI] [PMC free article] [PubMed] [Google Scholar]