Abstract
Escherichia coli RecA is a representative of proteins from the RecA family, which promote homologous pairing and strand exchange between double-stranded DNA and single-stranded DNA. These reactions are essential for homologous genetic recombination in various organisms. From NMR studies, we previously reported a novel deoxyribose-base stacking interaction between adjacent residues on the extended single-stranded DNA bound to RecA protein. In this study, we found that the same DNA structure was induced by the binding to Saccharomyces cerevisiae Rad51 protein, indicating that the unique DNA structure induced by the binding to RecA-homologs was conserved from prokaryotes to eukaryotes. On the basis of this structure, we have formulated the structure of duplex DNA within filaments formed by RecA protein and its homologs. Two types of molecular structures are presented. One is the duplex structure that has the N-type sugar pucker. Its helical pitch is ≈95 Å (18.6 bp/turn), corresponding to that of an active, or ATP-form of the RecA filament. The other is one that has the S-type sugar pucker. Its helical pitch is ≈64 Å (12.5 bp/turn), corresponding to that of an inactive, or ADP-form of the RecA filament. During this modeling, we found that the interconversion of sugar puckers between the N-type and the S-type rotates bases horizontally, while maintaining the deoxyribose-base stacking interaction. We propose that this base rotation enables base pair switching between double-stranded DNA and single-stranded DNA to take place, facilitating homologous pairing and strand exchange. A possible mechanism for strand exchange involving DNA rotation also is discussed.
Keywords: homologous recombination/Rad51 protein/homologous pairing/deoxyribose-base stacking
Since the structure of deoxyribose nucleic acid was first proposed by Watson and Crick (1, 2), it has been widely accepted that the way genetic information is stored, transmitted, and retrieved largely depends on the chemical properties of nucleic acids. RNA is characterized by its ability to form functional higher-order structures and by its reactivity, including self-cleavage and ligation of the sugar-phosphate backbone. The chemical activity of RNA is important for physiological reactions, especially in processes such as splicing and translation (3). In contrast to RNA, DNA has been recognized as a much less reactive entity with less structural diversity. Therefore, DNA has been characterized by its inactivity, which might be preferable for the faithful inheritance of genetic information. However, research in the past two decades has led to the increasing recognition that DNA potentially has a unique ability to search out homologous sequences in other DNA molecules, and this ability underlies homologous (genetic) recombination. Homologous recombination, which shuffles alleles between homologous chromosomes derived from two parents, is a general phenomenon in organisms with a DNA genome and is of primary importance for sexual reproduction in eukaryotes. In contrast, homologous recombination was observed less frequently in organisms with an RNA genome, e.g., RNA viruses (4). Homologous recombination also plays a central role in the repair of DNA damage.
In the various organisms with a DNA genome, proteins belonging to the RecA family play an essential role in homologous recombination. RecA protein, the first member of this family, is a 38-kDa protein encoded by the recA gene (5, 6) which is essential for homologous genetic recombination and DNA repair in Escherichia coli (7). RecA protein was first shown to promote the in vitro, ATP-dependent formation of joint molecules involving homologous single-stranded DNA and double-stranded DNA through “homologous pairing” and “strand exchange” (8–13). The joint molecules are general intermediates of homologous recombination and are pairs of DNA molecules connected by Watson–Crick hydrogen bonds (14).
How sequence homology is recognized between a single strand and fully duplex DNA remains an unanswered key question. In vitro studies revealed the following steps for the formation of the joint molecules by RecA protein: (i) in the presence of ATP (or its unhydrolyzable analogue, ATPγS), RecA protein cooperatively polymerizes along single-stranded DNA (15–17), resulting in the formation of “presynaptic filaments” in which the DNA is extended up to 1.5-fold compared with B-form DNA with the same sequence (18, 19); (ii) the presynaptic filament captures double-stranded DNA to search for homology to the single-stranded DNA present in the filament and upon homologous alignment of the DNA molecules a core heteroduplex is formed (the completion of homologous pairing; 15, 18, 20, 21); and (iii) the core heteroduplex is unidirectionally extended and stabilized by strand exchange (12, 22, 23; see also refs. 24–30 for review).
As described above, the presynaptic filament is the instrument for homologous pairing and strand exchange. Electron microscopic studies revealed common structural features between the nucleoprotein filament formed by RecA protein (19, 31–33) and that formed by other proteins of the RecA family including UvsX protein of coli phage T4 (34, 35) and Rad51 proteins of Saccharomyces cerevisiae (36) and Homo sapiens (37). Thus, it is very likely that a common mechanism mediates the catalysis of homologous pairing and strand exchange throughout the living kingdom.
Homologous pairing had been formally explained by either of two alternative models: (i) single-stranded DNA recognizes homology in double-stranded DNA by partially disrupting base pairs to make use of Watson–Crick interactions; or (ii) by the use of non-Watson–Crick interactions, single-stranded DNA pairs with double-stranded DNA whereas the latter retains its Watson–Crick base pairs. The former model first appeared to be supported by the observed ability of RecA protein to unwind double-stranded DNA (32, 38–40), but this interpretation was later challenged by the unfavorable observation that DNA that has been partially unwound by RecA protein remains base paired (41, 42). On the other hand, RecA protein was shown to form a three-stranded structure as an intermediate for the formation of the joint molecules (43–46). These observations favor the latter model. In the RecA reaction, single-stranded DNA might interact a priori initially with double-stranded DNA in either the major groove or the minor groove. Homologous alignment of single-stranded DNA and double-stranded DNA is potentially explained by triplex formation in which, by forming additional base pair specific hydrogen bonds, the single strand is placed in the major groove of the double-stranded DNA with the identical strands in a parallel orientation (47–50). Some reports described observations favorable to the initial interaction from the minor groove (51–56), but the advantage to homologous pairing has not been clarified to date.
ATP hydrolysis is necessary for extensive strand exchange especially to bypass heterologous blocks (57, 58), whereas homologous pairing does not require ATP hydrolysis and ATP can be replaced by an unhydrolyzable ATP-analog as a cofactor (59). Various biochemical studies indicated that homologous pairing and strand exchange can occur by using the nonhydrolyzable ATP analog, ATPγS (60, 61), or by a mutant RecA protein (RecA K72R) that exhibits >600-fold reduced level of an NTP hydrolysis (62, 63). Active RecA filaments formed in the presence of both ATP and DNA exhibit an extended structure with helical pitch of ≈95 Å (19, 31–33), whereas in the presence of ADP or the absence of nucleotide cofactors, RecA protein formed collapsed filaments with a helical pitch of ≈64 Å (64–68).
We recently reported a deoxyribose-base stacking interaction within extended single-stranded DNA bound to active RecA protein (69). To examine possible conservation of the DNA structure from prokaryotes to eukaryotes, we studied single-stranded DNA bound to yeast Rad51 protein by the same technique used in the previous structural study and found that Rad51 protein induced the same extended structure in single-stranded DNA upon binding. During further examination of the extended DNA by model building of duplex DNA, we found that the interconversion of sugar puckers induced horizontal base rotation in the extended structure. These findings suggest a mechanistic model for homologous pairing and strand exchange.
METHODS
NMR Spectroscopy.
Rad51 protein of S. cerevisiae was purified from an E. coli cell overexpressing the RAD51 gene on a plasmid as described (70) with a minor modification, which will be described elsewhere. Sample preparations and NMR experiments were carried out as described in our previous paper (69).
Structural Calculation and the Construction of DNA Models.
To investigate the structure of duplex DNA within recombination protein filaments, structures were first calculated on the basis of NMR studies of RecA-bound single-stranded DNA (69). A simulated annealing protocol was performed by embedding the interproton distances obtained from the NMR structure, using B-form DNA, d(TATATA), in Arnott configuration as a starting structure (71). The resulting duplex DNA structure had a helical pitch of ≈64 Å per turn, with the S-type sugar puckers. Then, we tried to construct a duplex DNA structure whose helical pitch was ≈95 Å per turn, corresponding to the active RecA filaments, and found that it required that the sugar puckers of the duplex should be, at least partly, in the N-type. These structural considerations led us to the idea of the base rotation mechanism driven by the conversion of sugar puckers.
To construct the duplex DNA model presented in Fig. 2, structures were generated by translational and rotational operations of standard coordinates of guanine:cytosine or adenine:thymidine base pairs in either B-form or A-form structure, and we connected bonds between sugar and phosphate of adjacent residues. Then, we minimized the energy of DNA structure by using the program x-plor (A. T. Brünger, Yale University, New Haven, CT) with parameters revised by Parkinson et al. (71, 72); first, base planes and furanose-rings were fixed to minimize the coordinates, then we repeated the minimization without any constraints. For triplex DNA structure, the N-type and S-type DNA structures were first connected, and then the structure was minimized as described above. Nucleotide residues of the replaced single-stranded DNA were placed without specific constraint. All calculations were carried out by using Silicon Graphics (Mountain View, CA) workstations.
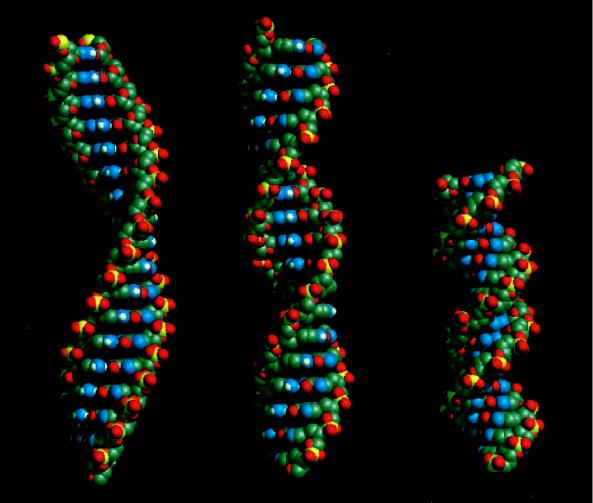
Figure 2.
Molecular models of the N-type (Left) and S-type (Center) duplex DNA structure within RecA protein filaments and B-form DNA (Right). The values of their pitches are set to 95 Å (18.6 bp/turn) for the N-type structure and 64 Å (12.5 bp/turn) for the S-type structure to satisfy those of active and inactive forms of RecA filaments, respectively. B-form DNA has a helical pitch of ≈36 Å (10.5 bp/turn). Each DNA molecule contains 18 base pairs. Note that both N-type and S-type duplex are extended by 1.5 times compared with B-form DNA but have different helical pitches. The N-type duplex DNA structure has a broad and open minor groove so that the single-stranded DNA can approach the duplex without severe steric hindrance.
RESULTS
Molecular Structure of Single-Stranded DNA Bound to the Yeast Rad51 Protein.
We have recently shown by NMR spectroscopy that the RecA-bound single-stranded DNA includes a deoxyribose-base stacking interaction in which a 2′-methylene moiety of a deoxyribose is placed above the base of the next residue, instead of following the normal stacking of adjacent bases (69). This structure explains well the 1.5-fold extension of DNA in RecA filaments compared with B-form DNA, which was previously observed by electron microscopic studies. Moreover, the extended structure includes little steric hindrance between adjacent residues, suggesting that the RecA-bound DNA allows for free movement of the bases. This process would be required during “base pair switching” from the original base pairs to the heteroduplex.
It would be an interesting question whether the unique extended structure is conserved in single-stranded DNA bound to eukaryotic RecA protein-homologs because various RecA homologs form very similar nucleoprotein filament around DNA and extend the DNA. To address this question, using the same technique applied to our previous DNA structural study, we analyzed single-stranded oligo-DNA bound to S. cerevisiae Rad51 protein. The S. cerevisiae Rad51 protein-bound DNA and E. coli RecA protein-bound DNA show similar transferred nuclear Overhauser effect (TRNOE) spectra (Fig. 1 a and b vs. Fig. 1 c and d). Unusually intense interresidue crosspeaks between H3′ and H8/H6 were observed, whereas weak, if any, interresidue crosspeaks between H1′ and H8/H6 were detected. We also observed relatively weak sequential H2′-H8/H6 and H2′′-H8/H6 NOEs of comparable intensity. These are common features to both spectra and a remarkable contrast to those expected for B-form or A-form DNA. They indicate that Rad51 protein-bound DNA has a similar extended structure as RecA protein-bound DNA. Thus, the unique extended DNA structure induced by the binding to RecA-homologs was conserved from prokaryotes to eukaryotes and therefore the structure would have fundamental importance.
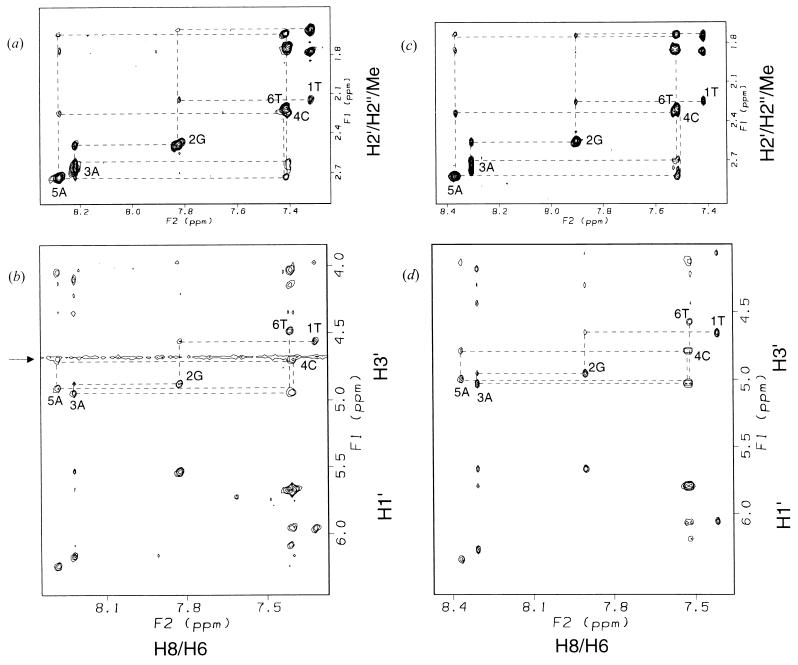
Figure 1.
TRNOE spectra of d(TGACAT) bound to S. cerevisiae Rad51 protein or E. coli RecA protein in the presence of ATPγS. (a and b) d(TGACAT) bound to Rad51 protein. The 0.71 mM d(TGACAT), 67 μM S. cerevisiae Rad51 protein, 0.71 mM ATPγS, 20 mM d11-Tris⋅Cl (pH* 7.1), and 6.7 mM MgCl2 in D2O at 303 K. (c and d) d(TGACAT) bound to RecA protein. The 1.1 mM d(TGACAT), 54 μM E. coli RecA protein, 1.1 mM ATPγS, 20 mM d11-Tris⋅Cl (pH* 7.1), and 6.7 mM MgCl2 and 150 mM NaCl in D2O at 310 K. Mixing time of both spectra is 200 msec. The dotted lines indicate sequential connectivities of d(TGACAT). Signals around 4.7 ppm (indicated by an arrow in b) are derived from residual water.
Molecular Structure of Duplex DNA in the RecA Filaments.
On the basis of the NMR solution to the structure of single-stranded DNA in the RecA filament, we constructed a model for duplex DNA in the filament because electron microscopic studies suggested that single-stranded DNA and double-stranded DNA complexed with RecA protein has a similar extended structure (19). To construct the molecular model, we made the following assumptions for the DNA structure, some of which are independently supported by biochemical or biophysical observations: (i) The duplex DNA includes the deoxyribose-base stacking interaction at each base step (69). (ii) Each DNA strand pairs through Watson–Crick type hydrogen bonds (41, 42). (iii) The base planes are perpendicular to the helical axis of the duplex DNA (42, 73). (iv) The distance between adjacent base pairs is 5.1 Å (31–33, 74). (v) The whole structure of duplex DNA is composed of a conformationally uniform building block of nucleotides.
First, we assumed that the sugar pucker of the deoxyribose was the S-type (near C2′-endo) conformation. We could obtain the structure of duplex DNA whose axial pitch was ≈64 Å (12.5 bp/turn; Fig. 2 Center), coincident with that of the inactive form of the RecA filament (64–68), but we were unable to build the structure that had the pitch of the active form of RecA filaments. On the other hand, the structure with an axial pitch of 95 Å (18.6 bp/turn) corresponding to the active form of the RecA filament (19, 31–33) was obtained when we assumed that the sugar pucker of deoxyribose was of the N-type (near C3′-endo) conformation (Fig. 2 Left).
Base Rotation by Interconversion of Sugar Puckers.
In homologous pairing and strand exchange, the RecA nucleoprotein filament, containing a single strand of DNA, catalyzes the transfer of a complementary strand from a homologous duplex molecule into the filament and subsequently the extrusion of the resulting unpaired strand. These processes involve a chain of conformational changes in DNA called base pair switching, which consist of the dissociation of base pairs of double-stranded DNA, movement of unpaired bases, and re-pairing to bases of the other complementary strand. In the RecA reactions, such base pair switching must occur efficiently to allow for a homology search between double-stranded DNA and single-stranded DNA and then to extend the formation of heteroduplex, all within a short time. Movement of unpaired bases in the reactions can occur in either of two ways: (i) by a rotational and translational motion of bases, in which the base plane moves roughly horizontal to the adjacent bases; or (ii) by a flip-flop motion of bases, in which the base plane rotates around a glycosyl bond between anti- and syn geometry. However, the flip–flop movement seems unlikely to be involved in these reactions because all of the exchanged base pairs must flip–flop back again to recover B-type structure.
Although we obtained well-defined NMR structures by structural calculations, the type of sugar puckers in the RecA-bound DNA was neither pure C2′-endo nor C3′-endo but something in between. In fact, the intensities of intra-residue crosspeaks of H3′-H8/H6 as well as H2′-H8/H6 were unexpectedly large in the 2-dimensional TRNOE spectra. One possible explanation for this result is that the sugar moieties in the RecA-bound DNA actually took on an intermediate pucker, but we also considered the possibility that RecA-bound DNA was in a dynamic state in which the S-type and N-type conformations of sugar puckers fluctuate in solution.
During the model-building study described in the previous section, we found that in the DNA structure that contains deoxyribose-base stacking, the interconversion of sugar puckers results in nearly horizontal rotation of base planes, without any severe steric hindrance (Fig. 3a). The conversion of a furanose ring from the N-type to S-type allows for the rotation of a base-pairing vector toward its minor groove. Moreover, because the deoxyribose-base stacking persists after the conversion from N-type to S-type, the extended structure also persists throughout the conformational change (Fig. 3b).
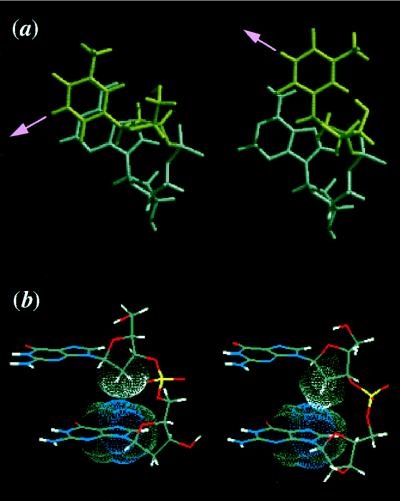
Figure 3.
Base rotation by interconversion of sugar puckers. (a) Top view of the base rotation caused by interconversion of the sugar puckers. The sugar pucker of the 5′-residue (T, top) is in the S-type (Left) and the N-type (Right), whereas that of the 3′-residue (A, bottom) is fixed in the S-type. Note that the hydrogen-bonding vector is rotated toward its major groove by the conversion from the S-type to the N-type. (b) Two types of deoxyribose-base stacking. All residues are in the S-type sugar pucker (Left) or the N-type (Right) sugar pucker.
Considering this rotation mechanism, we have constructed a molecular model of DNA triplex structure that demonstrates the base pair switching driven by the interconversion of sugar puckers. Fig. 4 indicates how base pairs of duplex DNA unpair and exchange their partners by this rotation mechanism. In Fig. 4a, the bottom three residues have the N-type sugar puckers and the top three residues have the S-type, while maintaining the extended structure with the deoxyribose-base stacking. Because the structure extended by the deoxyribose-base stacking includes little steric hindrance between adjacent residues (69), it presumably facilitates the reversible transition between these two states. Accordingly, we propose that this base rotation by interconversion of sugar puckers enables the switching of base pairs between double-stranded and single-stranded DNA, which is required both in the search for homology and in the formation of the heteroduplex DNA.
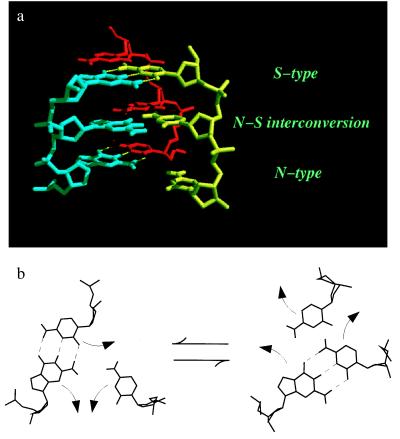
Figure 4.
A three-stranded DNA model for homology search and strand exchange considering the N–S interconversion of sugar puckers. (a) A molecular model of base pair switch between single- and double-stranded DNA. The bottom three residues are in the N-type and the top three residues are in the S-type. Note that the base pairing is altered by the conversion of sugar puckers between the N- and S-type. (b) Base rotation schemes for base pair switching against the interconversion of the sugar puckers. The bases are rotated toward the minor groove when the sugar puckers are converted from the N-type (Left) to the S-type (Right) and toward the major groove with the opposite (S-type to N-type) conversion.
DISCUSSION
Considering our previous structural determinations by NMR, we have constructed a molecular structure of duplex DNA in RecA filaments and obtained two types of duplex structures that are compatible with the helical parameters of active and inactive RecA filaments, respectively. One is the N-type structure with the N-type sugar pucker, whose helical pitch is ≈95 Å. The other is the S-type structure with the S-type sugar pucker, whose helical pitch is ≈64 Å. Both structures are extended by the deoxyribose-base-stacking interaction (69). Although our duplex DNA structures rest on experimental data (31–33, 41, 42, 69, 73, 74) and stereochemical arguments, they are insufficient for rigorous determination of the structure. Therefore, they need further experimental data to prove their validity. Nevertheless, we believe that our structure provides a reliable basis that can explain how single-stranded DNA recognizes homology in double-stranded DNA and how identical strands are subsequently exchanged.
It is interesting that the helical pitch values of the N-type and S-type duplex structure coincide with those of the active and inactive forms of RecA filaments, respectively. It seems reasonable to suppose that the N-type structure is the one within an active RecA filament because it agrees with the known helical parameters of duplex DNA within the active filament; where RecA protein unwinds, the duplex by 18.6 bp/turn and elongates it ≈1.5-times (31, 32, 39). In a previous model-building study, Zhurkin et al. (48) presented a triplex DNA structure in which sugar puckers were in the N-type but did not take into account the deoxyribose-base stacking revealed by our recent NMR study. A FTIR spectroscopic study also suggested that a triplex DNA in an active RecA-nucleoprotein filament had the N-type sugar conformation (75). The model proposed by Zhurkin et al. (48) postulated that all three bases were hydrogen bonded to each other, placing constraints on the formation of the triple helix. Such constraints, however, are not necessary for our model.
Our model building showed that the helical pitch of the S-type duplex structure corresponds to that of the inactive RecA filament. It seems reasonable to suppose that the N-type duplex in the active filament alters to the S-type duplex upon ATP hydrolysis of RecA protein. However, the binding affinity of an inactive filament to duplex DNA is very low, and consequently, no information for duplex DNA within an inactive filament is available. Therefore, the S-type duplex structure just provides a possible molecular model for a change induced in duplex DNA when RecA hydrolyzes ATP.
We showed that, in the deoxyribose-base-stacking structure, bases are rotated by the interconversion of sugar puckers while maintaining the extended DNA structure. This conversion process meets little steric hindrance between adjacent residues. (In contrast, it is theoretically shown that the conversion from the S-type to N-type is energetically restrained in B-form DNA because the 5′-side furanose ring hinders the movement of the 3′-side base moiety; ref. 76). Furthermore, because the energy barrier for the conversion of the sugar puckers is assumed to be sufficiently low (in the case of a purine mono ribonucleoside, it is estimated as ≈4.7 kcal/mole experimentally; ref. 77), the chemical equilibrium between the two states would be achieved. We propose that the DNA structure with the deoxyribose-base stacking has sufficient flexibility to allow the spontaneous conformational change required for base pair switching. This flexible property would be of advantage to the process of homologous pairing, because homologous pairing occurs very quickly without exogenous energy supply such as ATP hydrolysis (59–61). The base rotation through the conversion of sugar puckers might be driven by thermodynamic molecular motions and could progress easily by using the energy provided by external heat under physiological temperature.
Considering the idea of horizontal base rotation by interconversion of sugar puckers, we discuss here a possible mechanism by which homologous sequences between double-stranded DNA and single-stranded DNA are located (Fig. 4 a and b). First, we assumed that the reaction starts with the single-stranded and double-stranded DNA in the conformation associated with the N-type sugar pucker (Fig. 4a and b) because in that conformation the helical pitch of the duplex matches that of the activated filament where the reaction occurs. In the N-type structure, single-stranded DNA in the presynaptic filament should bind to the minor groove of duplex DNA for base pair switching because bases are rotated toward their minor grooves upon the conversion of the sugar puckers from the N-type to the S-type. When a base is rotated toward its minor groove (Fig. 4b, indicated by arrows) by this type of N-to-S conversion, it changes the base partner with which it pairs. Furthermore, the base pairing can return to its original configuration by reconversion of the sugar puckers from S-type to N-type. This base rotation can occur without ATP-hydrolysis through the flexible property. Successive trials for base pair exchanges by the interconversion of the sugar puckers would promote the search for homologous regions among the enormous number of base sequences.
If the bases in the newly formed combinations were homologously matched, the intermediate would be stabilized, at least temporarily, depending on the length of the homologous region. Sufficient homology between the single-stranded DNA and the double-stranded DNA would result in the formation of a core heteroduplex (homologous pairing).
If the tested base sequence was heterologous, the nascent intermediate would be unstable, and would immediately return to its former condition and finally dissociate. Here, the single-stranded DNA can slide relative to double-stranded DNA or can simply dissociate. The question of whether “a test-and-slide process” or “a test-and-dissociate process” takes place would depend on the DNA-binding properties of RecA protein, which we do not discuss here.
The question of whether the single-stranded DNA-RecA filament binds to the major or minor groove of duplex DNA is still controversial (48, 49, 51–56). In our description, we have assumed that it binds to the minor groove, but major groove binding is also possible with our model. If we were to assume the major groove-binding model, the reaction would start from the S-type DNA structure and end with the N-type structure.
Strand exchange is the result of a continuous series of base pair switches, and our model suggests that it is coupled with changes in the twist of the duplex between the N-type (≈18.6 bps/turn) and the S-type (≈12.5 bps/turn), which presumably cause DNA rotation around the helical axis. This filament rotation might play a role in facilitating uptake of an incoming DNA strand and displacement of an outgoing DNA strand. A role for the filament rotation and resulting torsional stress in strand exchange has been discussed in previous biochemical studies as a mechanism for unidirectional strand exchange and bypassing of heterologous inserts (78–82).
A number of RecA homologues have been found in various organisms, such as UvsX from bacteriophage T4 (83) and Rad51 from yeasts or mammals (70, 84). It has been demonstrated that several of them form helical filaments very similar to those produced by E. coli RecA protein (19, 31–37). Extended and unwound DNA has been observed commonly within those helical filaments (34, 36, 37, 85). In this study, we showed that the RecA-bound single-stranded DNA and the single-stranded DNA bound to S. cerevisiae Rad51 protein give similar TRNOE spectra, indicating that the Rad51-bound DNA has the same extended structure as RecA-bound DNA (Fig. 1). Thus, it is very likely that the DNA structure and molecular mechanisms that we described in this article are relevant to all of those similar filaments.
The extended structure with the deoxyribose-base stacking probably plays a critical role in the mechanisms of homologous recombination. The reactions of RecA protein would, at least partly, be attributed to the mobile abilities of the stacking structure of extended DNA in RecA filaments. Here, this deoxyribose-base joining can be regarded as a hinge to make it possible to rotate bases and alternate the combination of base pairs, where the methylene moiety acts as a pivot and the base as a saucer. The hydrophobic interaction through the deoxyribose-base stacking probably plays a role in stabilizing the whole structure while still allowing flexibility. These structural considerations lead us to suggest that along with its capacity to store and transmit genetic information, DNA is a precise molecular machine that is further suited to the subtle requirements of homologous recombination.
Acknowledgments
The authors would like to express their thanks to Dr. Charles M. Radding (Yale Medical School, New Haven, CT) for his critical reading of this manuscript and helpful suggestions, and to Drs. Judyth Sassoon and Liat Ben-Tovim for their proofreading of the manuscript. This study was partially supported by a grant for the “Biodesign Research Program” from The Institute of Physical and Chemical Research (RIKEN) and those from the Ministry of Education, Science and Culture, Japan. T.N. was formerly a Junior Research Associate and now holds a Special Post-doctoral Research fellowship at The Institute of Physical and Chemical Research (RIKEN).
ABBREVIATION
- TRNOE
transferred nuclear Overhauser effect
References
- 1.Watson J D, Crick F H C. Nature (London) 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Watson J D, Crick F H C. Nature (London) 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 3.Cech T R. Gene. 1988;73:259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- 4.Lai M M C. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horii T, Ogawa T, Ogawa H. Proc Natl Acad Sci USA. 1980;77:313–317. doi: 10.1073/pnas.77.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sancar A, Stachelek C, Konigsberg W, Rupp W D. Proc Natl Acad Sci USA. 1980;77:2611–2615. doi: 10.1073/pnas.77.5.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark A J, Margulies A D. Proc Natl Acad Sci USA. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McEntee K, Weinstock G M, Lehman I R. Proc Natl Acad Sci USA. 1979;76:2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata T, DasGupta C, Cunningham R P, Radding C M. Proc Natl Acad Sci USA. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassuto E, West S C, Mursalim J, Conlon S, Howard-Flanders P. Proc Natl Acad Sci USA. 1980;77:3962–3966. doi: 10.1073/pnas.77.7.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox M M, Lehman I R. Proc Natl Acad Sci USA. 1981;78:3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahn R, Cunningham R P, DasGupta C, Radding C M. Proc Natl Acad Sci USA. 1981;78:4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West S C, Cassuto E, Howard-Flanders P. Proc Natl Acad Sci USA. 1981;78:2100–2104. doi: 10.1073/pnas.78.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holliday R. Genet Res. 1964;5:282–304. [Google Scholar]
- 15.Shibata T, Cunningham R P, DasGupta C, Radding C M. Proc Natl Acad Sci USA. 1979;76:5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West S C, Cassuto E, Mursalim J, Howard-Flanders P. Proc Natl Acad Sci USA. 1980;77:2569–2573. doi: 10.1073/pnas.77.5.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahn R, Radding C M. J Biol Chem. 1984;259:7495–7503. [PubMed] [Google Scholar]
- 18.Howard-Flanders P, West S C, Stasiak A. Nature (London) 1984;309:215–220. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 19.Flory J, Tsang S S, Muniyappa K. Proc Natl Acad Sci USA. 1984;81:7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow S A, Radding C M. Proc Natl Acad Sci USA. 1985;82:5646–5650. doi: 10.1073/pnas.82.17.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonda D K, Shibata T, Radding C M. Biochemistry. 1985;24:413–420. doi: 10.1021/bi00323a026. [DOI] [PubMed] [Google Scholar]
- 22.Cox M M, Lehman I R. Proc Natl Acad Sci USA. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West S C, Cassuto E, Howard-Flanders P. Proc Natl Acad Sci USA. 1981;78:6149–6153. doi: 10.1073/pnas.78.10.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radding C M. Biochim Biophys Acta. 1989;1008:131–145. doi: 10.1016/0167-4781(80)90001-9. [DOI] [PubMed] [Google Scholar]
- 25.West S C. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- 26.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stasiak A, Egelman E H. Experientia. 1994;50:192–203. doi: 10.1007/BF01924002. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi M, Maraboeuf F, Norden B. Eur J Biochem. 1996;242:20–28. doi: 10.1111/j.1432-1033.1996.0020r.x. [DOI] [PubMed] [Google Scholar]
- 29.Kurumizaka H, Shibata T. J Biochem (Tokyo) 1996;119:216–223. doi: 10.1093/oxfordjournals.jbchem.a021224. [DOI] [PubMed] [Google Scholar]
- 30.Roca A I, Cox M M. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 31.Stasiak A, Di Capua E, Koller T. J Mol Biol. 1981;151:557–564. doi: 10.1016/0022-2836(81)90010-3. [DOI] [PubMed] [Google Scholar]
- 32.Dunn K, Chrysogelos S, Griffith J. Cell. 1982;28:757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- 33.Egelman E H, Stasiak A. J Mol Biol. 1986;191:677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- 34.Yu X, Egelman E H. J Mol Biol. 1993;232:1–4. doi: 10.1006/jmbi.1993.1363. [DOI] [PubMed] [Google Scholar]
- 35.Griffith J, Formosa T. J Biol Chem. 1985;260:4484–4491. [PubMed] [Google Scholar]
- 36.Ogawa T, Yu X, Shinohara A, Egelman E H. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 37.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunningham R P, Shibata T, DasGupta C, Radding C M. Nature (London) 1979;281:191–195. doi: 10.1038/281191a0. , (correction, 282, 4264). [DOI] [PubMed] [Google Scholar]
- 39.Stasiak A, Di Capua E. Nature (London) 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 40.Ohtani T, Shibata T, Iwabuchi M, Watabe H, Iino T, Ando T. Nature (London) 1982;299:86–89. doi: 10.1038/299086a0. [DOI] [PubMed] [Google Scholar]
- 41.Di Capua E, Muller B. EMBO J. 1987;6:2493–2498. doi: 10.1002/j.1460-2075.1987.tb02531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norden B, Elvingson C, Eriksson T, Kubista M, Sjöberg B, Takahashi M, Mortensen K. J Mol Biol. 1990;216:223–228. doi: 10.1016/S0022-2836(05)80311-0. [DOI] [PubMed] [Google Scholar]
- 43.Stasiak A, Stasiak A Z, Koller T. Cold Spring Harbor Symp Quant Biol. 1984;49:561–570. doi: 10.1101/sqb.1984.049.01.063. [DOI] [PubMed] [Google Scholar]
- 44.Hsieh P, Camerini-Otero C S, Camerini-Otero R D. Genes Dev. 1990;4:1951–1963. doi: 10.1101/gad.4.11.1951. [DOI] [PubMed] [Google Scholar]
- 45.Rao B J, Jwang B, Radding C M. J Mol Biol. 1990;213:789–809. doi: 10.1016/S0022-2836(05)80264-5. [DOI] [PubMed] [Google Scholar]
- 46.Umlauf S W, Cox M M, Inman R B. J Biol Chem. 1990;265:16898–16912. [PubMed] [Google Scholar]
- 47.Lacks S. Genetics. 1966;53:207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhurkin V B, Raghunathan G, Ulyanov N B, Camerini-Otero R D, Jernigan R L. J Mol Biol. 1994;239:181–200. doi: 10.1006/jmbi.1994.1362. [DOI] [PubMed] [Google Scholar]
- 49.Kim M G, Zhurkin V B, Jernigan R L, Camerini-Otero R D. J Mol Biol. 1995;247:874–889. doi: 10.1006/jmbi.1994.0187. [DOI] [PubMed] [Google Scholar]
- 50.Van Meervelt L, Vlieghe D, Dautant A, Gallois B, Precigoux G, Kennard O. Nature (London) 1995;374:742–744. doi: 10.1038/374742a0. [DOI] [PubMed] [Google Scholar]
- 51.Kumar K A, Muniyappa K. J Biol Chem. 1992;267:24824–24832. [PubMed] [Google Scholar]
- 52.Jain S K, Inman R B, Cox M M. J Biol Chem. 1992;267:4215–4222. [PubMed] [Google Scholar]
- 53.Chiu S K, Rao B J, Story R M, Radding C M. Biochemistry. 1993;32:13146–13155. doi: 10.1021/bi00211a025. [DOI] [PubMed] [Google Scholar]
- 54.Baliga R, Singleton J W, Dervan P B. Proc Natl Acad Sci USA. 1995;92:10393–10397. doi: 10.1073/pnas.92.22.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Podyminogin M A, Meyer R B, Gamper H B. Biochemistry. 1995;34:13098–13108. doi: 10.1021/bi00040a022. [DOI] [PubMed] [Google Scholar]
- 56.Zhou X H, Adzuma K. Biochemistry. 1997;36:4650–4661. doi: 10.1021/bi9630063. [DOI] [PubMed] [Google Scholar]
- 57.Rosselli W, Stasiak A. EMBO J. 1991;10:4391–4396. doi: 10.1002/j.1460-2075.1991.tb05017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J I, Cox M M, Inman R B. J Biol Chem. 1992;267:16438–16443. [PubMed] [Google Scholar]
- 59.Honigberg S M, Gonda D K, Flory J, Radding C M. J Biol Chem. 1985;260:11845–11851. [PubMed] [Google Scholar]
- 60.Menetski J P, Bear D G, Kowalczykowski S C. Proc Natl Acad Sci USA. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosselli W, Stasiak A. J Mol Biol. 1990;216:335–352. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- 62.Rehrauer W M, Kowalczykowski S C. J Biol Chem. 1993;268:1292–1297. [PubMed] [Google Scholar]
- 63.Shan Q, Cox M M, Inman R B. J Biol Chem. 1996;271:5712–5724. doi: 10.1074/jbc.271.10.5712. [DOI] [PubMed] [Google Scholar]
- 64.Koller, T., DiCapua, E. & Stasiak, A. (1983) ed. Cozzarelli, N. R. (Liss, New York), pp. 723–729.
- 65.Williams R C, Spengler S J. J Mol Biol. 1986;187:109–118. doi: 10.1016/0022-2836(86)90410-9. [DOI] [PubMed] [Google Scholar]
- 66.Heuser J, Griffith J. J Mol Biol. 1989;210:473–484. doi: 10.1016/0022-2836(89)90124-1. [DOI] [PubMed] [Google Scholar]
- 67.Yu X, Egelman E H. J Mol Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- 68.Ruigrok R W H, Bohrmann B, Hewat E, Engel A, Kellenberger E, Dicapua E. EMBO J. 1993;12:9–16. doi: 10.1002/j.1460-2075.1993.tb05626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nishinaka T, Ito Y, Yokoyama S, Shibata T. Proc Natl Acad Sci USA. 1997;94:6623–6628. doi: 10.1073/pnas.94.13.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 71.Brunger A T. x-plor Manual. New Haven, CT: Yale Univ. Press; 1987. , Version 3.1. [Google Scholar]
- 72.Parkinson G, Vojtechovsky J, Clowney L, Brünger A T, Berman H M. Acta Crystallogr D. 1996;52:57–64. doi: 10.1107/S0907444995011115. [DOI] [PubMed] [Google Scholar]
- 73.Norden B, Elvingson C, Kubista M, Sjoberg B, Ryberg H, Ryberg M, Mortensen K, Takahashi M. J Mol Biol. 1992;226:1175–1191. doi: 10.1016/0022-2836(92)91060-3. [DOI] [PubMed] [Google Scholar]
- 74.Di Capua E, Engel A, Stasiak A, Koller T. J Mol Biol. 1982;157:87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- 75.Dagneaux G, Porumb H, Liquier J, Takahashi M, Taillandier E. J Biomol Struct Dyn. 1995;13:465–470. doi: 10.1080/07391102.1995.10508856. [DOI] [PubMed] [Google Scholar]
- 76.Zhurkin V B, Gorin A A, Charakhchyan A A, Ulyanov N B. In: Sequence-Dependent Variability of A and B-DNA. Beveridge D L, Lavery R, editors. Schenectady, NY: Adenine; 1990. pp. 411–431. [Google Scholar]
- 77.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. [Google Scholar]
- 78.Cox M M, Lehman I R. Annu Rev Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- 79.Honigberg S M, Radding C M. Cell. 1988;54:525–532. doi: 10.1016/0092-8674(88)90074-8. [DOI] [PubMed] [Google Scholar]
- 80.Jwang B, Radding C M. Proc Natl Acad Sci USA. 1992;89:7596–7600. doi: 10.1073/pnas.89.16.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bedale W A, Cox M M. J Biol Chem. 1996;271:5725–5732. doi: 10.1074/jbc.271.10.5725. [DOI] [PubMed] [Google Scholar]
- 82.Cox M M. Trends Biochem Sci. 1994;19:217–222. doi: 10.1016/0968-0004(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 83.Fujisawa H, Yonesaki T, Minagawa T. Nucleic Acid Res. 1985;13:7473–7481. doi: 10.1093/nar/13.20.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 85.Harris L D, Griffith J. J Biol Chem. 1987;262:9285–9292. [PubMed] [Google Scholar]