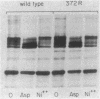
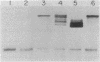
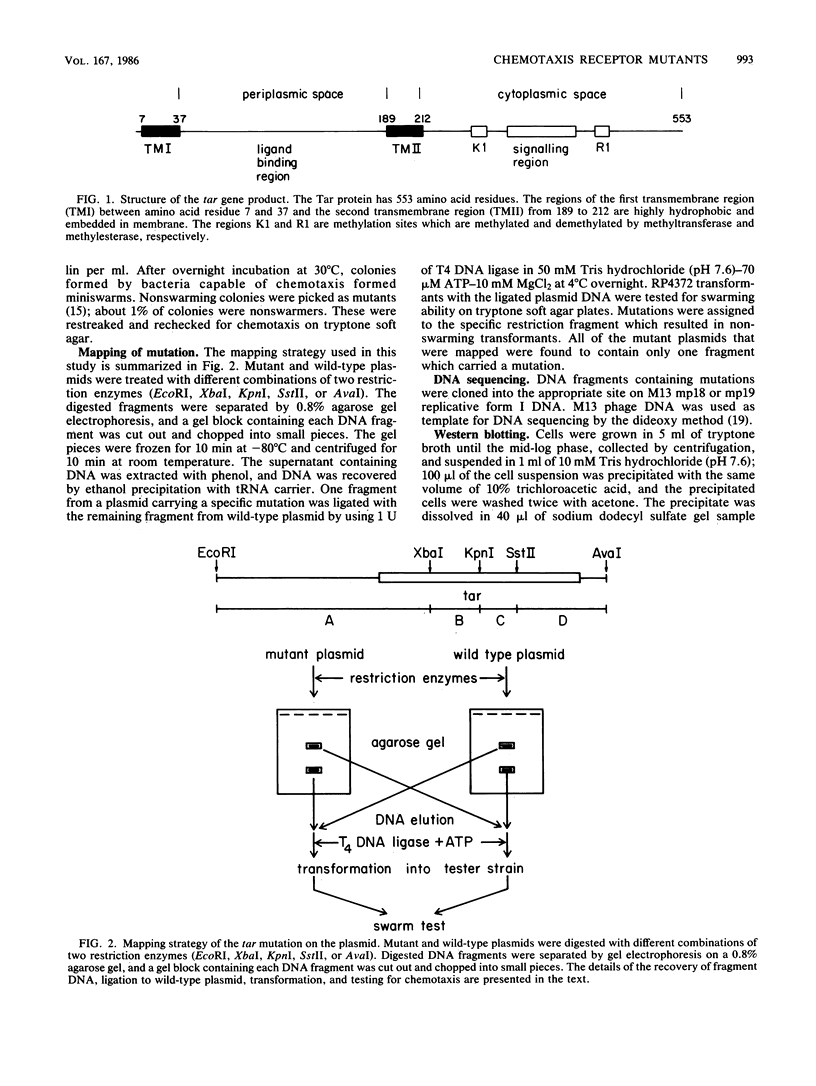
Abstract
Hydroxylamine mutagenesis was used to alter the tar gene that encodes the transmembrane Tar protein required for chemotaxis. Mutants defective in chemotaxis were selected, and the mutation was characterized by DNA sequencing. Two classes of mutations were found: nonsense and missense. The nonsense mutations were distributed throughout the gene, while the missense mutations were found to cluster in a region that includes 185 amino acids at the C-terminal end of the Tar protein. Partial characterization of mutant phenotypes suggested that some are completely defective in signaling while responding to attractants and repellents by differential methylation. Other mutants are undermethylated and constantly tumble, while yet another class of mutants is overmethylated and biased toward constant swimming with little or no tumbling. These mutants will be useful in experiments designed to understand the mechanism of chemotaxis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Block S. M., Segall J. E., Berg H. C. Impulse responses in bacterial chemotaxis. Cell. 1982 Nov;31(1):215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Callahan A. M., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: cheD mutations affect the structure and function of the Tsr transducer. J Bacteriol. 1985 Jan;161(1):96–104. doi: 10.1128/jb.161.1.96-104.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedblom M. L., Adler J. Chemotactic response of Escherichia coli to chemically synthesized amino acids. J Bacteriol. 1983 Sep;155(3):1463–1466. doi: 10.1128/jb.155.3.1463-1466.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Smith H. R., Anderson E. S. Mutagenesis of plasmid DNA with hydroxylamine: isolation of mutants of multi-copy plasmids. Mol Gen Genet. 1976 Apr 23;145(1):101–108. doi: 10.1007/BF00331564. [DOI] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Aberrant regulation of methylesterase activity in cheD chemotaxis mutants of Escherichia coli. J Bacteriol. 1985 Jan;161(1):105–112. doi: 10.1128/jb.161.1.105-112.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Sensory adaptation in bacterial chemotaxis: regulation of demethylation. J Bacteriol. 1985 Sep;163(3):983–990. doi: 10.1128/jb.163.3.983-990.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J Biol Chem. 1984 Oct 10;259(19):11828–11835. [PubMed] [Google Scholar]
- Krikos A., Conley M. P., Boyd A., Berg H. C., Simon M. I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ordal G. W. Bacterial chemotaxis: biochemistry of behavior in a single cell. Crit Rev Microbiol. 1985;12(2):95–130. doi: 10.3109/10408418509104426. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S. Novel mutations affecting a signaling component for chemotaxis of Escherichia coli. J Bacteriol. 1980 Jun;142(3):953–961. doi: 10.1128/jb.142.3.953-961.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J Bacteriol. 1976 May;126(2):758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Zanolari B. Sensory transduction in Escherichia coli: regulation of the demethylation rate by the CheA protein. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5061–5065. doi: 10.1073/pnas.81.16.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J., Kersulis G., Koshland D. E., Jr Neither methylating nor demethylating enzymes are required for bacterial chemotaxis. Cell. 1985 Sep;42(2):683–690. doi: 10.1016/0092-8674(85)90125-4. [DOI] [PubMed] [Google Scholar]