Abstract
Eukaryotic initiation factor (eIF) 4GI is a component of the cap-binding protein complex eIF4F, which is required for cap-dependent translation. Infection of cells by poliovirus results in a precipitous decline of host cell protein synthesis, which is preceded by the cleavage of eIF4GI. Cleavage of eIF4GI results in the inhibition of cap-dependent translation. Poliovirus translation is not affected by eIF4GI cleavage, however, because poliovirus mRNA is translated by a cap-independent mechanism. Cleavage of eIF4GI alone cannot explain the shutoff of host protein synthesis, because after infection in the presence of inhibitors of virus replication, eIF4GI is cleaved, yet host protein synthesis is only partially inhibited. Here we show that eIF4GII, a recently discovered functional homolog of eIF4GI, is more resistant to poliovirus-mediated cleavage than eIF4GI, and that its proteolysis is concomitant with the shutoff of host cell protein synthesis. Moreover, infection with poliovirus in the presence of inhibitors of virus replication resulted in efficient cleavage of eIF4GI, but only partial proteolysis of eIF4GII. Thus, cleavage of both eIF4GI and eIF4GII appears to be required for the shutoff of host protein synthesis after poliovirus infection. These results explain several earlier reports documenting the lack of correlation between eIF4GI cleavage and inhibition of cellular mRNA translation after poliovirus infection.
The dramatic shutoff of host protein synthesis after poliovirus infection has long been considered a prime example of translational control in eukaryotes. The current model to explain the mechanism of shutoff is based on the difference between the 5′ end of poliovirus mRNA (and other picornaviruses) and cellular mRNAs. While cellular mRNAs possess a cap structure (m7GpppX, where X is any nucleotide) at their 5′ end (1), poliovirus and other picornaviruses lack a cap structure (2, 3). Consequently, translation of cellular mRNAs generally proceeds by a cap-dependent mechanism, whereas translation of picornavirus RNAs proceeds by a cap-independent mechanism, in which ribosomes bind directly to an internal ribosome entry site in the 5′ untranslated region of the RNA (4–8).
Cap-dependent translation is mediated by the eukaryotic initiation factor (eIF) 4F, a three-subunit complex that binds to the cap structure (9). The three subunits are: (i) eIF4E, which interacts directly with the cap structure (10–12); (ii) eIF4A, an ATPase, which in conjunction with another initiation factor, eIF4B, exhibits RNA helicase activity (13); and (iii) eIF4G (formerly p220; ref. 14), which plays an important role in the assembly of the mRNA-ribosome initiation complex. eIF4G binds directly to several initiation factors, eIF3 (which interacts with the ribosome), eIF4E, and eIF4A (15, 16). We recently have cloned and characterized a novel human homolog of eIF4G, which we termed eIF4GII (17), while the original eIF4G (18) was renamed eIF4GI. eIF4GII is 46% identical to eIF4GI, binds eIF4E, eIF3, and eIF4A, and functionally complements eIF4GI to a significant extent (17).
The initial clue to explain the mechanism of poliovirus-induced shutoff of host protein synthesis was obtained by Etchison et al. (19). By using antisera against the 220-kDa component of eIF4F they demonstrated that the inhibition of host protein synthesis is preceded in vivo by the specific cleavage of eIF4G. This cleavage results in the production of two fragments: an amino-terminal polypeptide containing the eIF4E binding site (15, 16) and a carboxyl-terminal fragment that contains the binding sites for eIF3 and eIF4A (16, 20). As a result of this cleavage, cap-dependent translation is severely impaired. However, cap-independent translation of poliovirus RNA (and several other picornaviruses) requires only the carboxyl-terminal eIF4G fragment, as the amino-terminal fragment complexed with eIF4E is dispensable for their translation (21). The mediator of eIF4G cleavage for poliovirus and several other (but not all) picornaviruses is the viral protease 2A, which cleaves the poliovirus precursor polypeptide between P1 and 2Apro (reviewed in ref. 22). Recombinant 2Apro proteins of poliovirus, coxsackie, and rhinovirus cleave eIF4G in vitro to generate two fragments with sizes similar to those generated in vivo (23, 24). Thus, it was suggested that cleavage of eIF4G in vivo is accomplished directly by 2Apro (24). However, it also has been suggested that cleavage in poliovirus-infected cells occurs by 2Apro-mediated activation of an endogenous protease (25, 26).
Although cleavage of eIF4G can reasonably explain the shutoff phenomenon, there are many findings that are inconsistent with such a model. First, there is a lag of about 20–30 min, and even longer, between eIF4G cleavage and shutoff of host protein synthesis after poliovirus infection (19, 27, 28). More worrisome, however, are the observations that under certain conditions of infection, eIF4G is completely cleaved, whereas inhibition of host mRNA translation is only moderate. For example, Bonneau and Sonenberg (27) showed that when infection of poliovirus is carried out in the presence of guanidine-HCl or 3-methyl quercetin, inhibitors of viral replication (29, 30), cleavage of eIF4G occurs with kinetics similar to that observed upon infection in the absence of these drugs. However, under these conditions, host protein synthesis is inhibited only moderately (27). Based on these results it was concluded that a second event in addition to eIF4G cleavage must occur to effect a complete shutoff of host protein synthesis (27). Similar results were obtained by Carrasco’s group when infection was conducted in the presence of guanidine or other inhibitors of virus replication, such as monensin and nigericin (28, 31). However, these authors interpreted their results to indicate that eIF4G cleavage is irrelevant to the shutoff phenomenon (28, 31). In other experiments, Davies et al. (32) reported that expression of poliovirus 2Apro in COS-1 cells resulted in complete cleavage of eIF4G, whereas protein synthesis was inhibited only by 3-fold.
eIF4GII, the recently described homolog of eIF4GI (17), is also cleaved in vivo in poliovirus-infected cells and in vitro by rhinovirus 2Apro. However, a comparison of the kinetics of cleavage of eIF4GI and eIF4GII by 2Apro was not reported (17). The discovery of eIF4GII raised the intriguing possibility that its cleavage kinetics could differ from eIF4GI and might thus better explain the shutoff of host protein synthesis after poliovirus infection.
MATERIALS AND METHODS
Materials.
Materials were obtained from the following sources: chemiluminescence system, Amersham; nitrocellulose membrane, Schleicher & Schuell; [35S]methionine (1,000 Ci/mmol) and En3Hance, DuPont/NEN; guanidine-HCl, BioShop Canada (Ontario); monensin, Sigma; and protease inhibitor cocktail, Boehringer Mannheim.
Virus Infections, Drug Treatments, and Metabolic Labeling.
The Mahoney strain of poliovirus type 1 was used to infect HeLa R19 cells grown in DMEM containing 10% fetal bovine serum. Cells at ≈80% confluency were infected in serum-free medium at a multiplicity of infection (MOI) of 20, 50, or 100 plaque-forming units (pfu) per cell as indicated in the figure legends. All of the experiments also were performed with a MOI of 100 pfu/cell, with no substantial difference in the outcome. After virus adsorption at room temperature for 30 min, cells were further incubated at 37°C.
For the drug treatments, after virus adsorption, cell monolayers on 10-cm Petri dishes were immediately incubated at 37°C in 5 ml of methionine-free DMEM (GIBCO) in the presence or absence of guanidine-HCl (1.5 mM) or monensin (50 μM).
At the times indicated in the figure legends cells were incubated for 30 min with [35S]methionine (10 μCi/ml; 1 Ci = 37 GBq) at 37°C, before harvesting and processing for autoradiography and Western blotting. The results presented are a representative of experiments repeated at least twice with similar results.
Antibodies and Western Blotting.
A polyclonal antibody against an N-terminal fragment of eIF4GII was described previously (17). Polyclonal antibodies against N-terminal and C-terminal peptides of eIF4GI (a kind gift from L. Carrasco, Universidad Autónoma de Madrid) were described previously (33).
For harvesting, monolayers were washed once with PBS. Cells were scraped on ice into buffer (20 mM Hepes-KOH, pH 7.6/150 mM KCl/1 mM DTT/1 mM EDTA/protease inhibitor cocktail) and lysed by three freeze-thaw cycles. Cell debris was pelleted by centrifugation, and protein concentration in the supernatant was measured by the Bio-Rad assay. Proteins were denatured with an equal volume of 2× Laemmli sample buffer and boiled for 2 min. Proteins were subjected to SDS/PAGE and blotted onto 0.45 μm nitrocellulose membranes. The membranes were blocked overnight at 4°C in 5% skim milk and probed with the various antibodies for 4–5 h at 4°C in 10 mM Tris⋅HCl, pH 8.0/150 mM NaCl/0.5% Tween 20/5% skim milk. The blots were washed and subsequently incubated in the same buffer with donkey anti-rabbit Ig-horseradish peroxidase (Amersham) at a 1:5,000 dilution, for 45 min at room temperature. After extensive washing, the blots were developed with the Renaissance chemiluminescence system (ECL; Amersham). The relative intensities of the bands were quantified by using a BioImage Densitometer (Milligen/Biosearch).
The results shown are a representative of experiments repeated at least twice with similar results.
RESULTS
Temporal Correlation Between Inhibition of Host Protein Synthesis by Poliovirus and Cleavage of eIF4GI and eIF4GII.
Infection of HeLa cells by the poliovirus type I Mahoney led to an expected shutoff of host cell protein synthesis (Fig. 1A). Inhibition of host protein synthesis was detectable as early as 2 h and 15 min after infection (Fig. 1A, lane 7), and was almost complete by 2 h and 45 min after infection (Fig. 1 A, lane 9). To investigate the correlation between the inhibition of host protein synthesis and the cleavage of eIF4GI and eIF4GII, extracts from infected cells were analyzed by Western blotting. Cleavage of eIF4GI, as judged by the disappearance of the full-length protein and the appearance of the previously described cleavage products (19), was detected by 1 h after infection (Fig. 1B, compare lanes 1 and 4) and was almost complete by 1 h and 30 min (Fig. 1B, lane 6). This finding confirms earlier reports (19, 27, 28) showing that cleavage of eIF4GI precedes the shutoff of host protein synthesis (compare lane 6 of Fig. 1B and lane 5 of Fig. 1A). In contrast, eIF4GII cleavage was slower than that for eIF4GI (Fig. 1C). At 2 h postinfection, 30% of eIF4GII remained intact, whereas between 2 h and 2 h and 15 min postinfection eIF4GII was almost completely cleaved (Fig. 1C, compare lanes 8 and 9); note that the signal obtained for the cleavage products of eIF4GII is stronger than that detected for full-length eIF4GII. This could be caused by inefficient transfer of the full-length protein to the nitrocellulose membrane in this experiment). The quantitation of the cleavage of eIF4GI and eIF4GII is shown in Fig. 1D. Thus, clearly, eIF4GII proteolysis lags behind that of eIF4GI, and is concomitant with the complete inhibition of host cell protein synthesis upon poliovirus infection. These data suggest that cleavage of both eIF4GI and eIF4GII are required for the complete shutoff of host protein synthesis.
Figure 1.
Shutoff of host protein synthesis and cleavage of eIF4GI and eIF4GII upon poliovirus infection. (A) Pattern of protein synthesis. HeLa cells were mock-infected or infected with poliovirus (100 pfu per cell), and labeled for 30 min with [35S]methionine at the indicated times after infection, and equal amounts of cytoplasmic protein extracts (5 μg) were subjected to SDS/15% PAGE. The arrows indicate viral capsid proteins. (B and C) Analysis of eIF4GI and eIF4GII cleavage. Proteins (40 μg) of samples as in A were resolved by SDS/6% PAGE, transferred to nitrocellulose paper, and incubated with a polyclonal antibody against the N-terminal fragment of eIF4GI (B) or eIF4GII (C) as described in Materials and Methods. Positions of molecular mass standards (in kDa) are shown. (D) Quantitative analysis of the results in B and C. The amount of intact eIF4GI and eIF4GII present in each lane is presented as a percentage of that of uncleaved eIF4G (lane 1, no change was observed in the amount of eIF4G protein in mock-infected cells). p.i., postinfection.
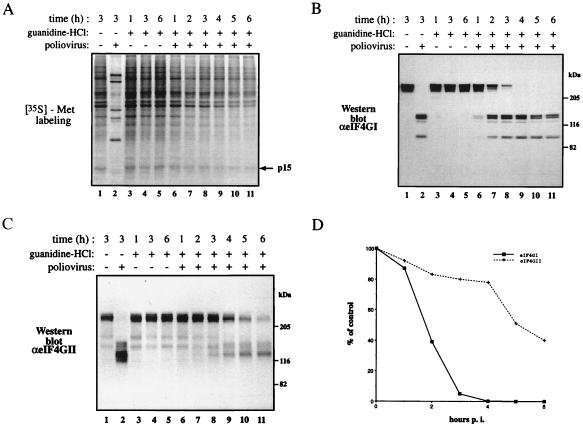
Guanidine-HCl Inhibits the Poliovirus-Mediated Shutoff of Host Cell Protein Synthesis and Proteolysis of eIF4GII.
To further investigate the hypothesis that cleavage of eIF4GII correlates with complete inhibition of host protein synthesis, we performed poliovirus infection in the presence of guanidine-HCl. This drug prevents poliovirus replication by inhibiting the function of the poliovirus 2C protein, which plays an essential role in viral replication (34). Because viral replication is inhibited, synthesis of poliovirus proteins, including 2Apro, is diminished. Poliovirus infection in the presence of guanidine-HCl results in a partial shutoff of host protein synthesis (27, 28). However, eIF4GI is completely cleaved under these conditions, suggesting that an additional event is required for the complete shutoff of host protein synthesis (27). To explore the possibility that cleavage of eIF4GII is the postulated second event, we examined its cleavage in the presence of guanidine-HCl. We performed these experiments with multiplicity of infections (MOIs) of 20 and 100 pfu/cell (the data with 20 pfu/cell are presented; similar results were obtained with a MOI of 100 pfu/cell). Poliovirus infection in the presence of guanidine resulted in a partial inhibition of host protein synthesis [80% inhibition for the cellular protein p15 even after 6 h of infection (similar values were obtained for other cellular proteins); Fig. 2A, compare lanes 5 and 11]. Under these conditions, eIF4GI was almost completely cleaved at 4 h after infection (Fig. 2B, compare lanes 3 and 9). However, in contrast, eIF4GII was almost intact at 4 h after infection (Fig. 2C, lane 9), and 60% cleaved at 6 h after infection (Fig. 2C, lane 11). The quantitation of the cleavage of eIF4GI and eIF4GII is shown in Fig. 2D. These results are consistent with the requirement of eIF4GII cleavage for the complete inhibition of host protein synthesis.
Figure 2.
Effects of guanidine-HCl on the shutoff of host protein synthesis in poliovirus-infected HeLa cells and cleavage of eIF4GI and eIF4GII. (A) HeLa cells were infected with poliovirus (20 pfu per cell) in the presence or absence of guanidine-HCl (1.5 mM) and labeled for 30 min with [35S]methionine at the indicated times after infection. Equal amounts of cytoplasmic extract (5 μg) were subjected to SDS/15% PAGE. The arrow indicates an arbitrarily chosen cellular protein (p15) that was used to measure the inhibition of host protein synthesis. (B and C) Analysis of eIF4GI and eIF4GII cleavage. Proteins (40 μg) of samples as in A were resolved by SDS/6% PAGE, transferred to nitrocellulose paper, and incubated with a mixture of polyclonal antibodies against the N- and C-terminal fragments of eIF4GI (B) or the N-terminal fragment of eIF4GII (C) as described in Materials and Methods. Positions of molecular mass standards (in kDa) are shown. (D) Quantitative analysis of the results in B and C. The amount of intact eIF4GI and eIF4GII present in each lane is presented as a percentage of that of uncleaved eIF4G (lane 3, no significant change was observed in the amount of eIF4G proteins in mock-infected cells). p.i., postinfection.
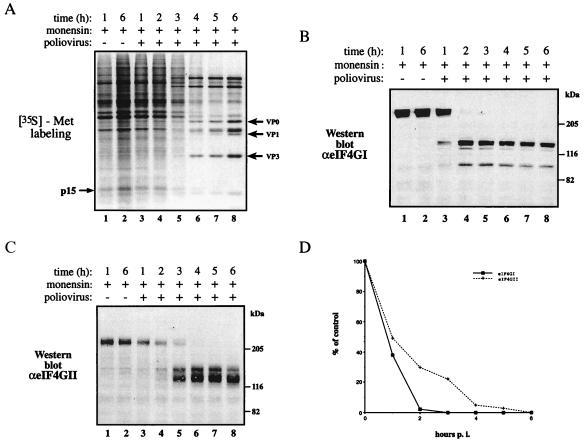
Effects of Monensin on the Poliovirus-Induced Shutoff of Host Translation and Cleavage of eIF4GI and eIF4GII.
To further study the differential sensitivity of eIF4GI vs. eIF4GII to cleavage, we wanted to use, in addition to guanidine-HCl, another inhibitor of poliovirus replication. Monensin is a carboxylic ionophore that exchanges Na+ for H+ and interferes with cellular vesicular traffic (35). Previous studies showed that monensin slowed the synthesis of poliovirus proteins and prevented the inhibition of cellular mRNA translation (31). When monensin was present during infection, inhibition of host protein synthesis was delayed as compared with infection in its absence (31). In the present study, at 3 h postinfection cellular protein synthesis was 29% of wild-type levels as determined by densitometric analysis of p15 (Fig. 3A, compare lanes 1 and 5). In the presence of monensin, cleavage of eIF4GI proceeded with similar kinetics to that observed in the absence of monensin (compare Fig. 3B to Figs. 2B and 1B). In contrast, cleavage of eIF4GII was impaired by monensin, and at 3 h, 22% of eIF4GII remained intact (Fig. 3C, lane 5), whereas eIF4GI was completely cleaved (Fig. 3B, lane 5). Thus, cleavage of eIF4GII coincides with the complete inhibition of host protein synthesis (compare lane 7 of Fig. 3A with lane 7 in Fig. 3C). The quantitation of the cleavage of eIF4GI and eIF4GII is shown in Fig. 3D.
Figure 3.
Effects of monensin on the shutoff of host protein synthesis and cleavage of eIF4GI and eIF4GII after poliovirus infection. (A) HeLa cells were infected with poliovirus (50 pfu per cell) in the presence or absence of monensin (50 μM) and labeled for 30 min with [35S]methionine at the indicated times after infection. Equal amounts of cytoplasmic protein (5 μg) were subjected to SDS/15% PAGE. The arrows on the right indicate viral capsid proteins. The arrow on the left indicates an arbitrarily chosen cellular protein (p15). (B and C) Analysis of eIF4GI and eIF4GII cleavage. Proteins (40 μg) of samples as in A were resolved by SDS/6% PAGE, transferred to nitrocellulose paper, and incubated with a mixture of polyclonal antibodies against the N- and C-terminal fragments of eIF4GI (B) or the N-terminal fragment of eIF4GII (C) as described in Materials and Methods. Positions of molecular mass standards (in kDa) are shown. (D) Quantitative analysis of the results in B and C. The amount of intact eIF4GI and eIF4GII present in each lane is presented as a percentage of that of the uncleaved eIF4G (lane 1, no significant change was observed in the amount of eIF4Gs in mock-infected cells). p.i., postinfection.
In Vitro Cleavage Analysis.
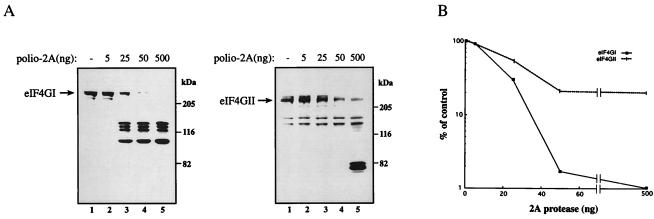
The previous results suggest that eIF4GII may be a poorer substrate for 2Apro as compared to eIF4GI. To further study the substrate properties of eIF4GII, we performed a dose–response analysis of the cleavage of the two eIF4G proteins by purified poliovirus 2Apro. HeLa cytoplasmic extract was incubated with increasing amounts of 2Apro (a generous gift of R. Banerjee, University of California, Los Angeles; ref. 36). The cleavage of eIF4GI and eIF4GII subsequently was monitored by immunoblotting with anti-eIF4GI and anti-eIF4GII polyclonal antibodies. eIF4GI was stable when incubated with buffer alone (Fig. 4A, lane 1, Left), but was efficiently cleaved into its characteristic cleavage products when incubated with increasing amounts of poliovirus 2Apro (Fig. 4A, Left, lanes 2–5). Under these conditions, 25 ng of poliovirus 2Apro cleaved approximately 70% of the eIF4GI (Fig. 4B). Furthermore, the migration of the cleavage products resembled the pattern observed in vivo from poliovirus-infected HeLa cells in the presence and in the absence of inhibitors of virus replication (Figs. 1B, 2B, and 3B).
Figure 4.
In vitro kinetics of cleavage of eIF4Gs by purified poliovirus 2Apro. (A) HeLa cytoplasmic extracts were prepared as described in Materials and Methods with the exception that the protease inhibitor mixture (Boehringer) was omitted from the lysis buffer. The extract (14 μl; 100 μg) was incubated in the absence (lanes 1) or presence (lanes 2–5) of the indicated amounts of poliovirus 2Apro in buffer containing 20 mM Hepes-KOH (pH 7.6), 150 mM KCl, 1 mM DTT, 1 mM EDTA for 30 min at 30°C, in a final volume of 35 μl. An equal volume of Laemmli buffer was added to stop the reaction. Samples were resolved by SDS/8% PAGE, and proteins were transferred onto nitrocellulose membranes, blotted, and incubated with a mixture of polyclonal antibodies against the N- and C-terminal fragments of eIF4GI (Left, 18-μl reaction in each lane), or the N-terminal fragment of eIF4GII (Right, 36-μl reaction in each lane). Positions of molecular mass standards (in kDa) are shown. (B) Quantitative analysis of the results in A. The amount of intact eIF4GI and eIF4GII present in each lane is presented as a percentage of that of input eIF4G (lanes 1). The experiment was repeated three times with similar results.
In contrast to the efficient cleavage of eIF4GI in the HeLa cytoplasmic extract, eIF4GII was a relatively poor substrate for cleavage by poliovirus 2Apro (Fig. 4A, Right). Although buffer alone had no effect on eIF4GII (Fig. 4A, Right, lane 1), even addition of 500 ng of poliovirus 2Apro did not cause complete cleavage of eIF4GII as 14% of the protein remained intact (Fig. 4A, Right, lane 5; the cleavage products derived from the amino terminus of eIF4GII displayed a faster mobility than those observed in vivo in poliovirus-infected HeLa cells, Figs. 1C and 3C; we do not have an explanation for this aberrant mobility). Thus, in vitro, as it is in vivo, eIF4GII appears to be more resistant than eIF4GI to cleavage by poliovirus 2Apro. These conclusions were further reinforced by showing that poliovirus 2Apro cleaved recombinant eIF4GI much more efficiently than recombinant eIF4GII (data not shown).
DISCUSSION
In this paper we describe experiments that appear to solve a long-standing puzzle regarding the mechanism of the shutoff of host protein synthesis after poliovirus infection. Although eIF4GI is invariably cleaved upon infection with poliovirus and several other picornaviruses, numerous earlier works reported a lack of correlation between eIF4GI cleavage and the shutoff of host protein synthesis in poliovirus-infected cells (27, 28, 31, 32). However, the earlier apparent contradictions, now may be explained by the relative resistance of eIF4GII to cleavage. Indeed, because eIF4GII serves as a functional homolog of eIF4GI in translation (17), virus-induced eIF4GII cleavage is required, in addition to cleavage of eIF4GI, for effective shutoff of cap-dependent translation.
The differential sensitivity of eIF4GI vs. eIF4GII to proteolytic cleavage might be important to the interpretation of other previous observations. For instance, injection of coxsackievirus B4 2Apro into Xenopus oocytes caused the complete cleavage of eIF4GI, yet protein synthesis decreased by only 35% (37). The authors concluded from these results that translation in Xenopus oocytes is cap independent, as it does not require intact eIF4GI. However, based on our current results the possibility that eIF4GII was not cleaved under the conditions used will need to be addressed.
What is the molecular basis for the differential susceptibility of eIF4GI vs. eIF4GII to poliovirus 2Apro? The cleavage site of eIF4GII by 2Apro has not been mapped, and there is only partial conservation between the 2Apro cleavage site in eIF4GI and the corresponding sequence in eIF4GII. The sequence for eIF4GI is TLSTR*GP, while that for eIF4GII is TPGGR*GV. The Gly residue at position P1′ (first position on the carboxyl-terminal side of the scissile bond) is essential for recognition of model eIF4GI peptides by 2A proteases in vitro (38). The P1′ Gly residue is conserved in eIF4GII. However, at position P4 (Leu in eIF4GI), which is also important for recognition by 2Apro (24, 38, 39), there is Pro in eIF4GII, which might not be compatible with efficient cleavage. Another explanation for the different protease sensitivities of the two eIF4G forms, which is not mutually exclusive, is the possibility that the two forms are present to different extents in various complexes (e.g., free or bound to eIF3 or eIF4E). It was demonstrated previously that eIF4GI is much more sensitive to cleavage by picornavirus 2Apro when in a complex with eIF4E than when uncomplexed (40, 41).
It would be of considerable interest to determine the sensitivity of eIF4GII cleavage by other picornavirus members and to correlate the shutoff of host protein synthesis engendered by these viruses with the cleavage of eIF4GI and eIF4GII. In one study it was shown that rhinovirus-14 cleaves eIF4GI very efficiently early (90 min) after infection (23). However, under these conditions, the shutoff of host protein synthesis occurs much later (23). We have found that eIF4GII is also more resistant to cleavage than eIF4GI in rhinovirus-14 infected cells (Y.V.S. and A.G., unpublished observations).
Other events, in addition to cleavage of eIF4GII, could accentuate the shutoff of host protein synthesis after poliovirus infection, such as the dephosphorylation of 4E-BP1, a specific inhibitor of eIF4E function (42), and membrane permeabilization (43). However, these events occur only late in infection and cannot explain the dramatic early inhibition of translation after infection.
The physiological significance of the differential sensitivity of eIF4GI vs. eIF4GII to cleavage by 2Apro is not known. Cleavage of eIF4G proteins has been reported recently, documenting the degradation of eIF4GI in reperfused rat ischemic brain neurons undergoing apoptosis (44). Also, recently, Lloyd et al. obtained data showing cleavage of eIF4GI during apoptosis (R. E. Lloyd, personal communication). If eIF4G cleavage plays an active role in apoptosis, then the differential cleavage sensitivities of eIF4GI vs. eIF4GII may have an important physiological outcome.
Acknowledgments
We thank A. Dasgupta and R. Banerjee for the poliovirus 2A protease, and L. Carrasco for anti-eIF4GI antibody. We are indebted to C. Lister for excellent technical assistance. We thank F. Callari and A. Kahwajian for help with computer graphics, and B. Raught, A.-C. Gingras, and S. Pyronnet for critical reading of the manuscript. This work was supported by a grant from the Medical Research Council of Canada to N.S., who is a Distinguished Scientist of the Medical Research Council of Canada and a Howard Hughes Institute International Scholar.
ABBREVIATIONS
- eIF
eukaryotic initiation factor
- pfu
plaque-forming unit
References
- 1.Shatkin A J. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Hewlett M J, Rose J K, Baltimore D. Proc Natl Acad Sci USA. 1976;73:327–330. doi: 10.1073/pnas.73.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nomoto A, Lee Y F, Wimmer E. Proc Natl Acad Sci USA. 1976;73:375–380. doi: 10.1073/pnas.73.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier J, Sonenberg N. Nature (London) 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 5.Jang S K, Krausslich H G, Nicklin M J H, Duke G M, Palmenberg A M, Wimmer E. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson R J, Howell M T, Kaminski A. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 7.Belsham G J, Sonenberg N. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrenfeld E. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 549–573. [Google Scholar]
- 9.Sonenberg N. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 245–269. [Google Scholar]
- 10.Sonenberg N, Rupprecht K M, Hecht S M, Shatkin A J. Proc Natl Acad Sci USA. 1979;76:4345–4349. doi: 10.1073/pnas.76.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcotrigiano J, Gingras A-C, Sonenberg N, Burley S K. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 12.Matsuo H, Li H, McGuire A M, Fletcher C M, Gingras A C, Sonenberg N, Wagner G. Nat Struct Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 13.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahara S M, Morgan M A, Shatkin A J. J Biol Chem. 1981;256:7691–7694. [PubMed] [Google Scholar]
- 15.Mader S, Lee H, Pause A, Sonenberg N. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 17.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan R, Rychlik W, Etchison D, Rhoads R E. J Biol Chem. 1992;267:23226–23231. [PubMed] [Google Scholar]
- 19.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 20.Imataka H, Sonenberg N. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohlmann T, Rau M, Pain V M, Morley S J. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan M D, Flint M. J Gen Virol. 1997;78:699–723. doi: 10.1099/0022-1317-78-4-699. [DOI] [PubMed] [Google Scholar]
- 23.Etchison D, Fout S. J Virol. 1985;54:634–638. doi: 10.1128/jvi.54.2.634-638.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamphear B J, Yan R, Yang F, Waters D, Liebig H D, Klump H, Kuechler E, Skern T, Rhoads R E. J Biol Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- 25.Lloyd R E, Toyoda H, Etchison D, Wimmer E, Ehrenfeld E. Virology. 1986;150:299–303. doi: 10.1016/0042-6822(86)90291-6. [DOI] [PubMed] [Google Scholar]
- 26.Bovee M L, Marissen W E, Zamora M, Lloyd R E. Virology. 1998;245:229–240. doi: 10.1006/viro.1998.9171. [DOI] [PubMed] [Google Scholar]
- 27.Bonneau A-M, Sonenberg N. J Virol. 1987;61:986–991. doi: 10.1128/jvi.61.4.986-991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez L, Carrasco L. Virology. 1992;189:178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 29.Bablanian R. Virology. 1972;47:255–259. doi: 10.1016/0042-6822(72)90260-7. [DOI] [PubMed] [Google Scholar]
- 30.Castrillo J L, Carrasco L. J Virol. 1987;61:3319–3321. doi: 10.1128/jvi.61.10.3319-3321.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irurzun A, Sanchezpalomino S, Novoa I, Carrasco L. J Virol. 1995;69:7453–7460. doi: 10.1128/jvi.69.12.7453-7460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies M V, Pelletier J, Meerovitch K, Sonenberg N, Kaufman R J. J Biol Chem. 1991;266:14714–14720. [PubMed] [Google Scholar]
- 33.Aldabe R, Feduchi E, Novoa I, Carrasco L. Biochem Biophys Res Commun. 1995;215:928–936. doi: 10.1006/bbrc.1995.2553. [DOI] [PubMed] [Google Scholar]
- 34.Pincus S E, Wimmer E. J Virol. 1986;60:793–796. doi: 10.1128/jvi.60.2.793-796.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tartakoff A M. Cell. 1983;32:1026–1028. doi: 10.1016/0092-8674(83)90286-6. [DOI] [PubMed] [Google Scholar]
- 36.Yalamanchili P, Banerjee R, Dasgupta A. J Virol. 1997;71:6881–6886. doi: 10.1128/jvi.71.9.6881-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keiper B D, Rhoads R E. Nucleic Acids Res. 1997;25:395–402. doi: 10.1093/nar/25.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sommergruber W, Ahorn H, Klump H, Seipelt J, Zoephel A, Fessl F, Krystek E, Blaas D, Kuechler E, Liebig H D, Skern T. Virology. 1994;198:741–745. doi: 10.1006/viro.1994.1089. [DOI] [PubMed] [Google Scholar]
- 39.Sommergruber W, Ahorn H, Zophel A, Maurer-Fogy I, Fessl F, Schnorrenberg G, Liebig H D, Blaas D, Kuechler E, Skern T. J Biol Chem. 1992;267:22639–22644. [PubMed] [Google Scholar]
- 40.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. J Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohlmann T, Pain V M, Wood W, Rau M, Morley S J. EMBO J. 1997;16:844–855. doi: 10.1093/emboj/16.4.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gingras A C, Svitkin Y, Belsham G J, Pause A, Sonenberg N. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrasco L. Adv Virus Res. 1995;45:61–112. doi: 10.1016/S0065-3527(08)60058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeGracia D J, Neumar R W, White B C, Krause G S. J Neurochem. 1996;67:2005–2012. doi: 10.1046/j.1471-4159.1996.67052005.x. [DOI] [PubMed] [Google Scholar]