Abstract
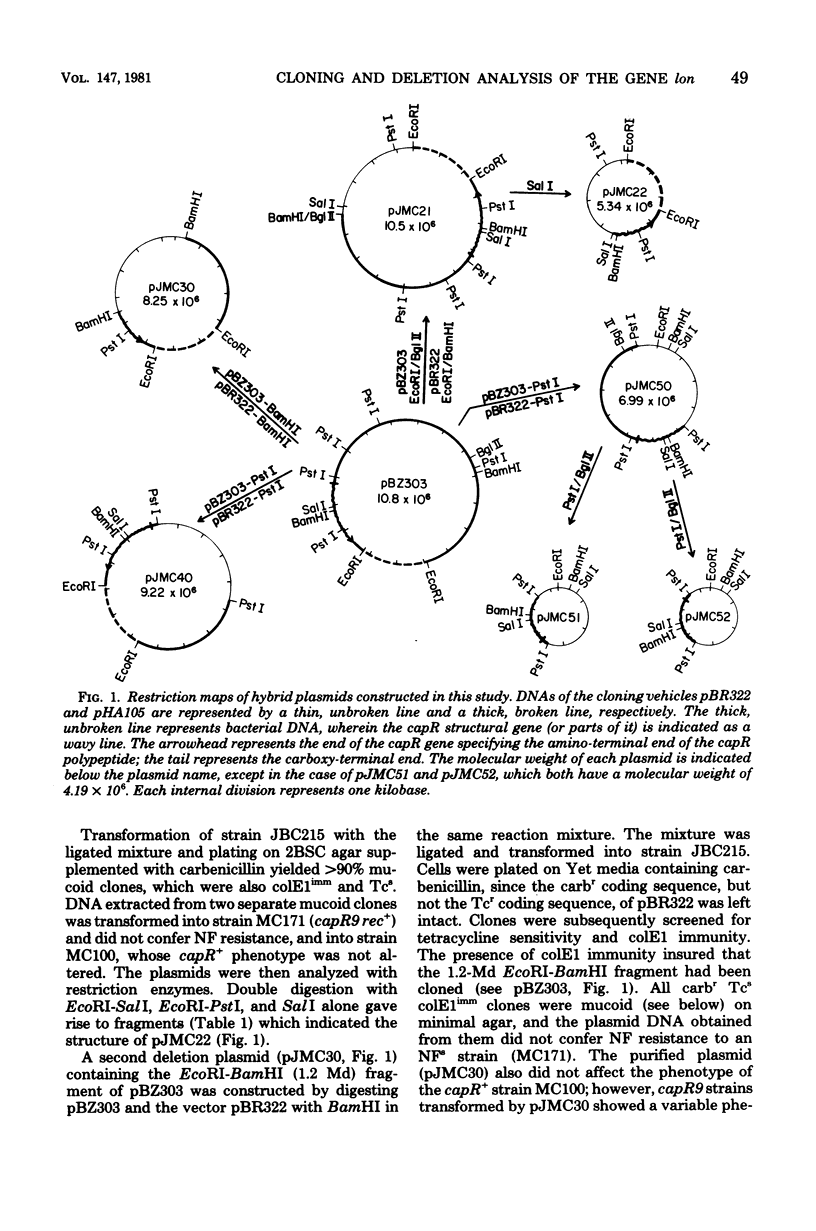
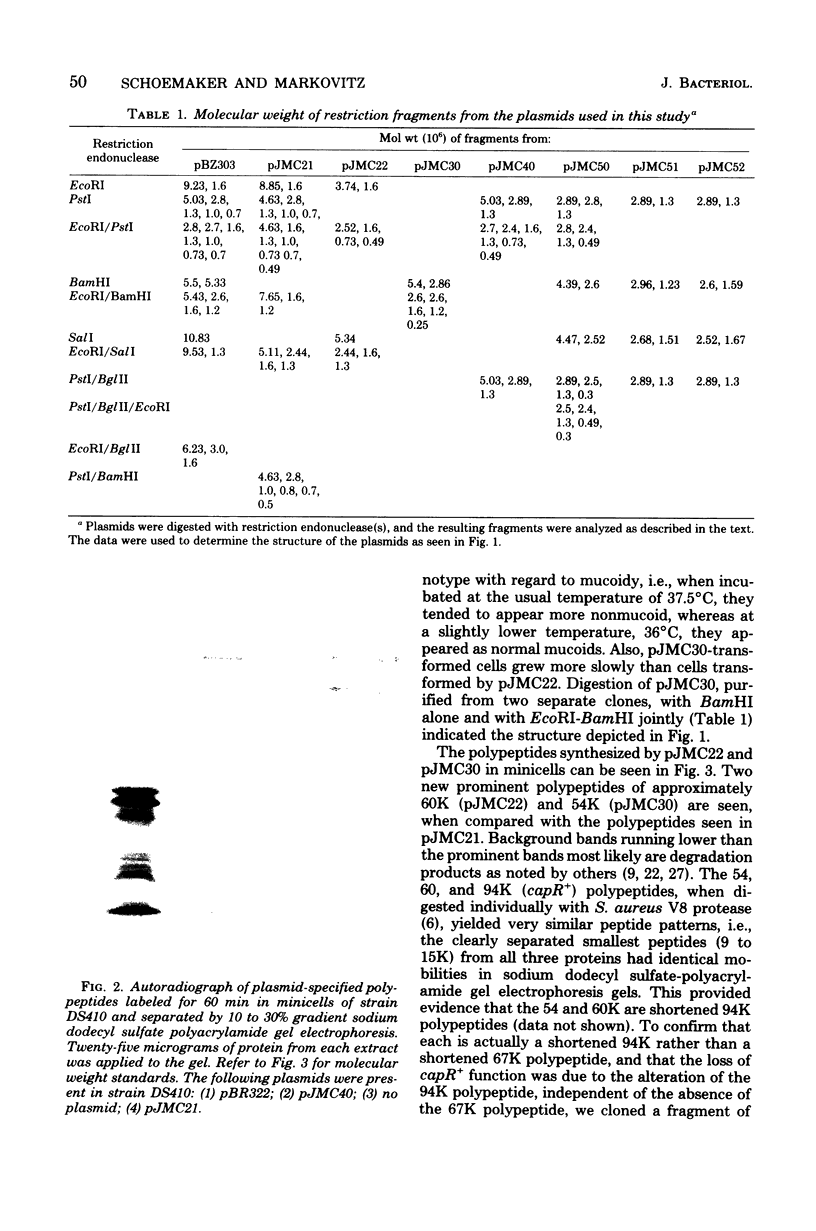
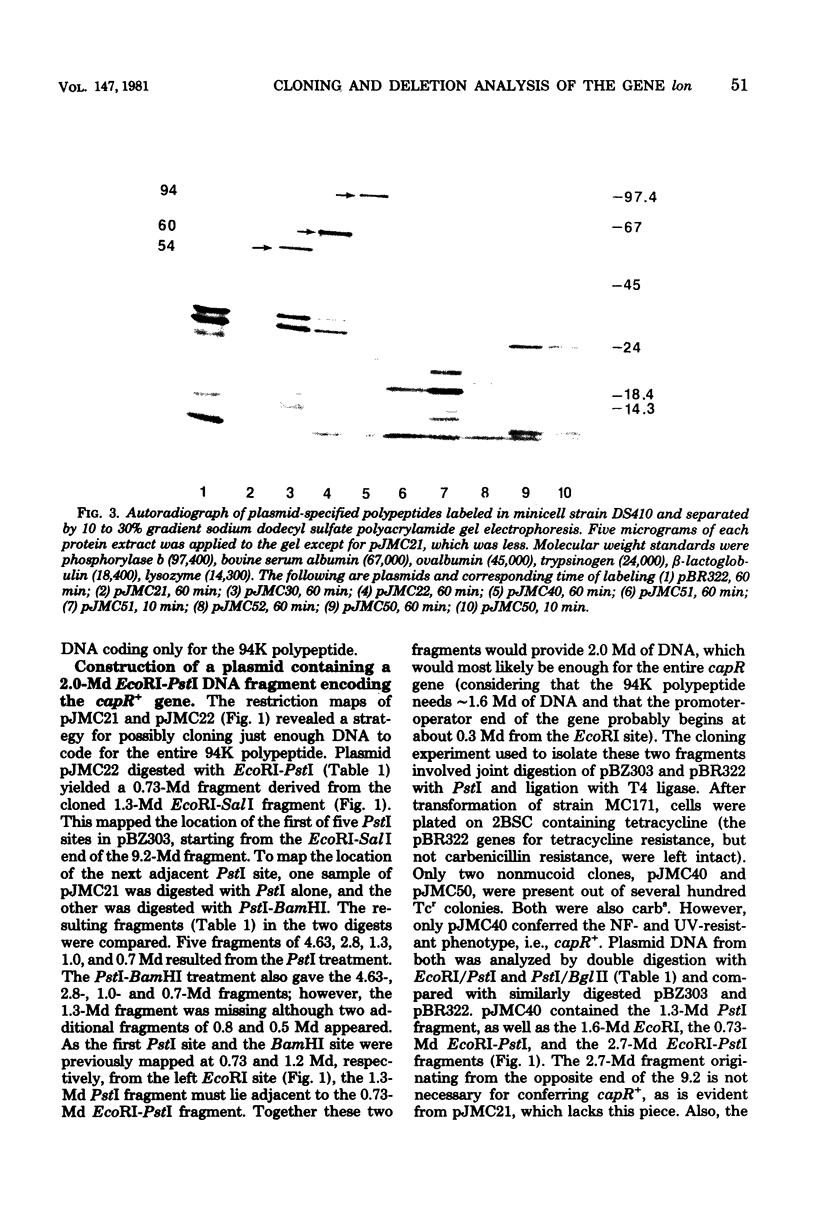
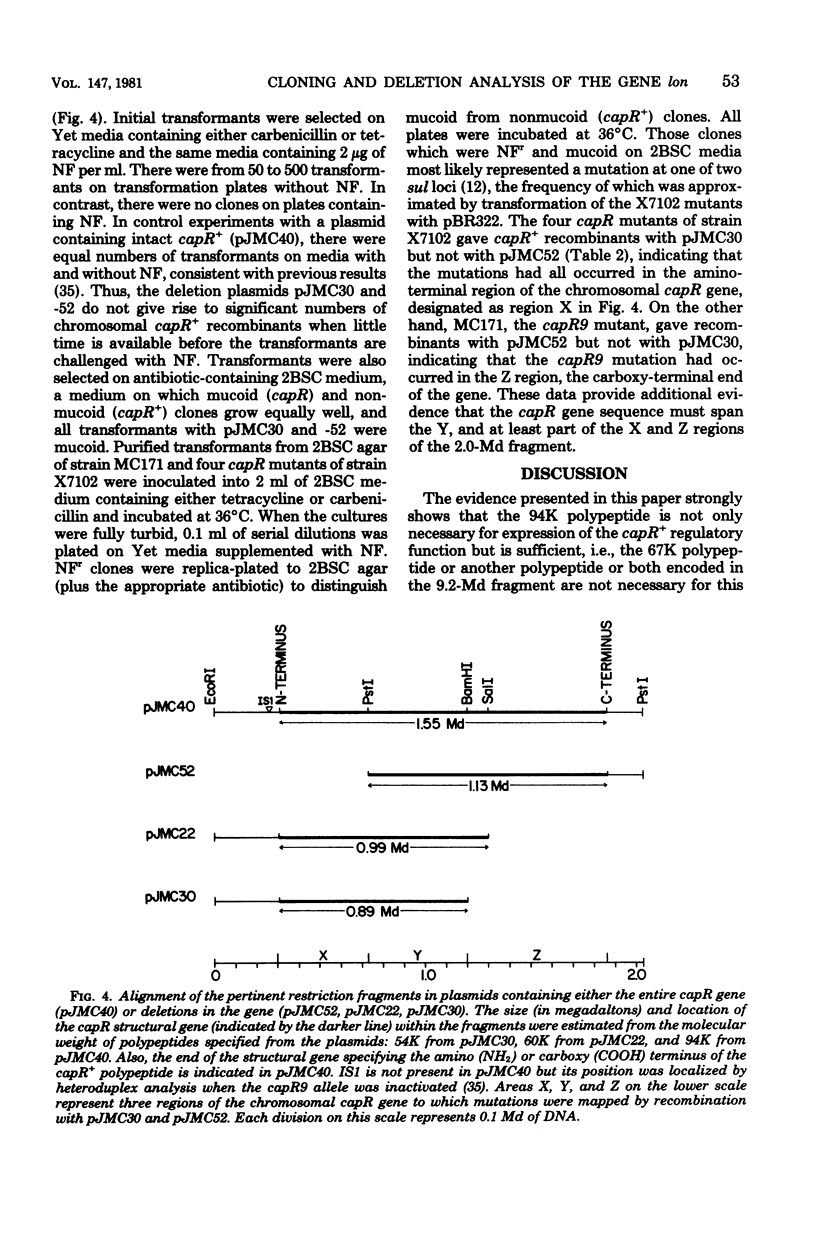
A mutation in the lon (capR) gene of Escherichia coli K-12 effects several phenotypic alterations in the mutant cell, such as overproduction of capsular polysaccharide and sensitivity to ultraviolet or ionizing radiation. A previously cloned 9.2-megadalton (Md) EcoRI fragment contained the capR+ gene and specified two polypeptides, 94 kilodaltons (K) and 67K. To provide evidence that the 94K polypeptide is the capR+ gene product, we constructed a capR+ plasmid pJMC40, having a 2.0-Md EcoRI-PstI fragment which codes only for the 94K polypeptide. Plasmids pJMC22 and pJMC30, having deletions of 0.7 and 0.8 Md, respectively, from one end of the 2.0-Md fragment, were also constructed. Each codes for a shortened stable polypeptide (from the 94K). Neither plasmid can confer the capR+ phenotype to capR mutants, confirming that the unaltered 94K polypeptide is the capR+ gene product. Plasmids pJMC51 and pJMC52 each have a deletion of 0.7 Md from the other end of the 2.0-Md fragment, differing only in the orientation of the remaining 1.3-Md fragment with respect to the cloning vehicle. They are nonfunctional with respect to capR+ and do not code for a common polypeptide from the 1.3-Md fragment. These data indicate that the fragments in pJMC22 and pJMC30, which both code for shortened 94K polypeptides, contain the promoter-operator region of the capR gene. The deletion plasmids were also used to map chromosomal capR mutations.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ADLER H. I., HARDIGREE A. A. ANALYSIS OF A GENE CONTROLLING CELL DIVISION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI. J Bacteriol. 1964 Mar;87:720–726. doi: 10.1128/jb.87.3.720-726.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni H., Berg P. E., Markovitz A. New mini-ColE1 as a molecular cloning vehicle. J Bacteriol. 1977 Jan;129(1):358–366. doi: 10.1128/jb.129.1.358-366.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni H., Markovitz A. Characterization of a mini ColE1 cloning vector. Plasmid. 1979 Apr;2(2):225–236. doi: 10.1016/0147-619x(79)90041-6. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bukhari A. I., Zipser D. Mutants of Escherichia coli with a defect in the degradation of nonsense fragments. Nat New Biol. 1973 Jun 20;243(129):238–241. doi: 10.1038/newbio243238a0. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Donch J., Greenberg J. Genetic analysis of lon mutants of strain K-12 of Escherichia coli. Mol Gen Genet. 1968;103(2):105–115. doi: 10.1007/BF00427138. [DOI] [PubMed] [Google Scholar]
- Frazer A. C., Curtiss R., 3rd Production, properties and utility of bacterial minicells. Curr Top Microbiol Immunol. 1975;69:1–84. doi: 10.1007/978-3-642-50112-8_1. [DOI] [PubMed] [Google Scholar]
- Gayda R. C., Avni H., Berg P. E., Markovitz A. Outer membrane protein a and other polypeptides regulate capsular polysaccharide synthesis in E. coli K-12. Mol Gen Genet. 1979 Oct 1;175(3):325–332. doi: 10.1007/BF00397232. [DOI] [PubMed] [Google Scholar]
- Gayda R. C., Markovitz A. Altered bacteriophage lambda expression in cell division mutants capR(lon) of Escherichia coli K-12. Mol Gen Genet. 1978 Feb 7;159(1):1–11. doi: 10.1007/BF00401741. [DOI] [PubMed] [Google Scholar]
- Gayda R. C., Markovitz A. Cloned DNA fragment specifying major outer membrane protein a in Escherichia coli K-12. J Bacteriol. 1978 Oct;136(1):369–380. doi: 10.1128/jb.136.1.369-380.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayda R. C., Yamamoto L. T., Markovitz A. Second-site mutations in capR (lon) strains of Escherichia coli K-12 that prevent radiation sensitivity and allow bacteriophage lambda to lysogenize. J Bacteriol. 1976 Sep;127(3):1208–1216. doi: 10.1128/jb.127.3.1208-1216.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Zipser D. Deg phenotype of Escherichia coli lon mutants. J Bacteriol. 1978 Feb;133(2):844–851. doi: 10.1128/jb.133.2.844-851.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby E. P., Ruff W. L., Goldthwait D. A. Cell division and prophage induction in Escherichia coli: effects of pantoyl lactone and various furan derivatives. J Bacteriol. 1972 Aug;111(2):447–453. doi: 10.1128/jb.111.2.447-453.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowit J. D., Goldberg A. L. Intermediate steps in the degradation of a specific abnormal protein in Escherichia coli. J Biol Chem. 1977 Dec 10;252(23):8350–8357. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. B. R factor proteins synthesized in Escherichia coli minicells: incorporation studies with different R factors and detection of deoxyribonucleic acid-binding proteins. J Bacteriol. 1974 Dec;120(3):1451–1463. doi: 10.1128/jb.120.3.1451-1463.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R. B., Tait R. C., Betlach M., Boyer H. W. Protein expression in E. coli minicells by recombinant plasmids. Cell. 1977 Mar;10(3):521–536. doi: 10.1016/0092-8674(77)90039-3. [DOI] [PubMed] [Google Scholar]
- Modrich P., Zabel D. EcoRI endonuclease. Physical and catalytic properties of the homogenous enzyme. J Biol Chem. 1976 Oct 10;251(19):5866–5874. [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N. Bacteriophage infection of minicells: a general method for identification of "in vivo" bacteriophage directed polypeptide biosynthesis. Mol Gen Genet. 1977 Dec 14;158(1):73–79. doi: 10.1007/BF00455121. [DOI] [PubMed] [Google Scholar]
- Reeve J. N. Selective expression of transduced or cloned DNA in minicells containing plasmid pKB280. Nature. 1978 Dec 14;276(5689):728–729. doi: 10.1038/276728a0. [DOI] [PubMed] [Google Scholar]
- Shineberg B., Zipser D. The ion gene and degradation of beta-galactosidase nonsense fragments. J Bacteriol. 1973 Dec;116(3):1469–1471. doi: 10.1128/jb.116.3.1469-1471.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T. Bacterial mutants defective in plasmid formation: requirement for the lon + allele. Proc Natl Acad Sci U S A. 1971 Jul;68(7):1469–1473. doi: 10.1073/pnas.68.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Zehnbauer B. A., Markovitz A. Cloning of gene lon (capR) of Escherichia coli K-12 and identification of polypeptides specified by the cloned deoxyribonucleic acid fragment. J Bacteriol. 1980 Aug;143(2):852–863. doi: 10.1128/jb.143.2.852-863.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]