Abstract
The binding of hemimethylated oriC to Escherichia coli membranes has been implicated in the prevention of premature reinitiation at newly replicated chromosomal origins in a reaction that involves the SeqA protein. We describe the resolution of the membrane-associated oriC-binding activity into two fractions, both of which are required for the high-affinity binding of hemimethylated oriC. The active component in one fraction is identified as SeqA. The active component of the second fraction is a previously undescribed protein factor, SeqB. The reconstituted system reproduced the salient characteristics of the membrane-associated binding activity, suggesting that the membrane-associated oriC-binding machinery of E. coli is likely to be a multiprotein system that includes the SeqA and SeqB proteins.
In Escherichia coli and most other bacterial species, chromosome replication is initiated at a specific point during the cell cycle (1). It is clear that a mechanism exists to prevent premature reinitiation until the proper time in the cell cycle is reached (2). Several factors have been proposed to play a role in the sequestration of newly replicated origins to prevent premature reinitiation during the eclipse period.
The methylation state of the chromosomal origin has been implicated both in the binding of oriC to membranes and in the prevention of premature reinitiation at newly replicated origins. In E. coli, adenine residues within chromosomal GATC sequences are methylated in a reaction catalyzed by Dam methylase. Because the methylation reactions occur after replication, adenine residues within GATC sequences in the newly synthesized strand remain unmethylated for a period of time. The oriC region contains a high density of GATC sites, and methylation of adenine residues at several of these sites is delayed significantly relative to the time of methylation at GATC sites elsewhere in the chromosome (3, 4). As a result, the oriC region of newly replicated chromosomes remains hemimethylated for about 30–40% of the cell cycle.
Evidence that hemimethylated oriC does not serve as an effective template for initiation of replication in vivo came from the finding of Russell and Zinder (5) that a fully methylated oriC plasmid was unable to replicate in a Dam− host. Under these conditions, the first round of replication in the Dam− host gives rise to hemimethylated oriC that is never fully methylated because of the defect in Dam methylase. It was suggested that the hemimethylated origin was refractory to reinitiation, thereby explaining the defect in plasmid replication. Ogden et al. (3) then showed that hemimethylated oriC DNA binds preferentially to membrane fractions in vitro. Flotation density gradient analysis showed that the oriC-binding activity was primarily localized to a unique membrane fraction (6).
Evidence that the association of hemimethylated oriC with the membrane plays a direct role in preventing initiation of replication came from the demonstration by Landoulsi et al. (7) that binding of hemimethylated oriC to an E. coli membrane preparation prevented the oriC fragment from acting as a template for DNA replication in an in vitro replication system. Taken together, these results support the idea that the membrane association of hemimethylated oriC plays a key role in the postreplication block of reinitiation that occurs during the normal cell cycle.
SeqA is a protein that is required for both the membrane binding of hemimethylated oriC in vitro and the refractory period to initiation that follows replication of the origin region in dam+ cells (8–10). This is consistent with the view that SeqA plays a role in sequestering the newly replicated origin by making it inaccessible to the replication machinery, and suggests that this may involve a SeqA-mediated oriC–membrane interaction. The nature of the putative membrane-binding site for hemimethylated oriC has not been defined and it is not known whether additional proteins in addition to SeqA may be involved. The possible role of another protein, HobH, that has been shown to bind to hemimethylated oriC in nuclease protection assays, also remains to be defined (11). We therefore have attempted to further characterize the component(s) of the membrane-associated oriC-binding machinery.
In this paper, we describe the dissociation of the E. coli membrane-associated oriC-binding activity into two fractions, both of which are required for the high-affinity binding of hemimethylated oriC. The active component of the first fraction is identified as SeqA. The active component of the second fraction includes a previously unrecognized protein, SeqB, that activates the oriC-binding activity of SeqA and is required for SeqA-oriC binding at low concentrations of SeqA. This suggests that the membrane-associated oriC-binding machinery of E. coli is a multiprotein complex that includes the SeqA and SeqB proteins.
MATERIALS AND METHODS
Preparation of oriC Substrates.
Fully methylated (+/+) and unmethylated (−/−) oriC fragments were prepared from the oriC plasmid pGO46, isolated from host strains UT481 (dam+) for +/+ oriC, and GM2929 (dam−) for −/− oriC, after overnight growth in LB medium at 37°C. The plasmids were digested with AvaI to obtain the 467-bp oriC fragments. Annealing of methylated and unmethylated strands was accomplished by heating a mixture of +/+ and −/− fragments [40 ng/μl each in STE buffer (12)] in a heating block successively for 10 min at 90°C, 10 min at 65°C, and 120 min at 28°C. The resulting mixture should contain approximately 50% hemimethylated oriC and 25% each of +/+ and −/− oriC. The +/+ and −/− fragments were removed by digestion with MboI and DpnI at approximate concentrations of 1.5 × 10−3 and 0.7 × 10−3 units/ng DNA, respectively, for 60 min at 37°C followed by 20 min at 65°C. Under these conditions the enzymes were specific for −/− and +/+ DNA, respectively, as verified independently for each experiment. The +/− oriC fragment was reisolated by preparative agarose gel electrophoresis, with a final yield of approximately 85% of the theoretical yield.
The +/−, +/+, and −/− oriC fragments were end labeled with [32P]CTP as previously described (6).
Gel Retardation Assay.
The sample to be assayed for binding activity (at the concentrations indicated in the figures) was mixed in buffer B [250 mM potassium glutamate/10 mM Pipes (pH 6.5)/2 mM EDTA] with 0.15 μg/ml sonicated calf thymus DNA. After incubation at room temperature for 15 min, 6 pM hemimethylated 32P-labeled oriC was added unless otherwise indicated. After standing at room temperature for 30 min, the reaction mixture (10 or 20 μl) was electrophoresed in a 4% or 7% polyacrylamide gel prepared in buffer B. The gel was then dried and analyzed by autoradiography. oriC-binding activity was expressed as the fraction of total 32P-labeled oriC that was recovered in the retarded band, as measured with a Packard IntantImager.
When fraction A and/or fraction B were assayed, the indicated amounts of each fraction [in 10 mM Pipes (pH 6.5)/2 mM EDTA/0.25 M potassium thiocyanate (KSCN)] were mixed and allowed to stand at 30°C for 20 min and then at room temperature for 20 min before addition to the reaction mixture. KSCN concentrations higher than 70 μM began to inhibit the binding reaction, and final thiocyanate concentrations were therefore adjusted to 50 μM.
Fractionation of oriC-Binding Components.
Membrane preparation. E. coli PC2 (dnaCts, dnaT, thyA, leuB, rpsL) was grown at 30°C to early stationary phase in LB medium supplemented with 0.025% thymine. For preparation of labeled membranes, the medium contained 10 μCi/ml [2-3H]glycerol (200 mCi/mmol). The washed cell pellet was suspended in 20 ml of 20% sucrose in buffer A [10 mM Hepes (pH 7.4)/5 mM EDTA] and disrupted in a French pressure cell (6). DNase (0.025 μg/μl) and MgCl2 (2 mM) were added. After 30 min at room temperature, unbroken cells were removed by centrifugation at 2,000 rpm for 10 min. The supernatant fraction (cell extract, 3 ml) was applied to the top of a step gradient (6 ml of 20% sucrose and 2 ml of 60% sucrose in buffer A) and spun for 3.5 hr at 4°C in a Beckman SW41 rotor (16). The material at the 20%/60% sucrose interface (membrane) was collected, suspended in 12 ml of buffer A, and pelleted by centrifugation in a SW41 rotor at 40,0000 rpm for 3.5 hr at 4°C. The final pellet was suspended in 2–3 ml of 50 mM Pipes (pH 6.5)/2 mM EDTA.
KSCN extraction.
Five-tenths milliliter of the membrane suspension, containing approximately 4 mg of protein, was added to 0.5 ml of 1 M KSCN/20 mM Pipes (pH 6.5). After standing for 60 min at room temperature, the sample was centrifuged for 90 min at 4°C in a Beckman TL100 rotor and the supernatant and pellet (KSCN pellet) fractions were collected. The supernatant fraction was diluted with 4 volumes of 0.19 M KSCN/20 mM Pipes (pH 6.5) to give the KSCN extract.
Preparation of fractions A and B.
The KSCN extract was dialyzed overnight at 4°C against 250 ml of 10 mM Pipes (pH 6.5)/2 mM EDTA, and the cloudy suspension that formed within the dialysis bag was centrifuged in a TL100 rotor at 4°C for 90 min. The supernatant was called fraction B. The precipitate was washed once in the same Pipes/EDTA buffer and recentrifuged. The final pellet (fraction A) was solubilized by suspension in 0.5 M KSCN/10 mM Pipes (pH 6.5)/2 mM EDTA. Fractions A and B were each adjusted to 0.255 M KSCN in a final volume of 2.2 ml so that 0.55 μl of each fraction corresponded to the amounts obtained from 1 μl of the KSCN extract.
Analytic Procedures.
Immunoblot analysis was performed after SDS gel electrophoresis in 12% polyacrylamide gels (13) using a polyclonal rabbit anti-SeqA antibody preparation.
Materials.
Purified SeqA (14) and anti-SeqA antibody were kind gifts from J. Epstein and N. Kleckner (Harvard University, Boston) and from E. Boye (Institute of Cancer Research, Norweigan Radium Hospital, Oslo), respectively.
RESULTS
Solubilization of Membrane-Associated oriC-Binding Activity.
As previously described (3, 6, 9), a crude E. coli membrane fraction was capable of binding hemimethylated 32P-labeled oriC. The membrane preparation showed significant oriC-binding activity at concentrations of 0.1 μg/ml of membrane protein or lower as measured in a gel retardation assay (Fig. 1a). This corresponds to a SeqA concentration of approximately 1–1.5 nM in the binding assay mixture based on quantitative immunoblot analysis of the membrane sample (data not shown). At this concentration, purified SeqA fails to show detectable binding of hemimethylated oriC (see Fig. 5 and ref. 9).
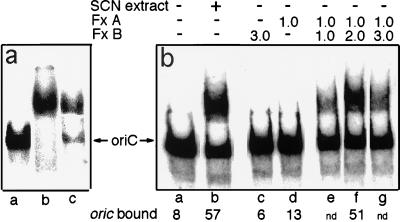
Figure 1.
Reconstitution of membrane-associated oriC-binding activity. (a) The gel retardation assay mixture (20 μl) contained either no additional components (lane a), membrane (2 μg, lane b), or KSCN extract (0.4 μg, lane c), representing the amount extracted from 2 μg of membrane. The arrow indicates the position of hemimethylated oriC in the absence of other components. (b) The gel retardation mixtures contained aliquots of: lane b, KSCN extract (1 μl); lane c, fraction B (1.65 μl); lane d, fraction A (0.55 μl); and lanes e–g, mixtures of fraction A (0.55 μl) and fraction B prepared as described in Materials and Methods; the amounts of fraction B in reactions e, f, and g were 0.55, 1.1 ,and 1.65 μl, respectively. The amounts of the binding components are expressed relative to the equivalent concentration of the KSCN extract (e.g., 1.0 corresponds to the amount of fraction A or B that was obtained from the amount of KSCN extract that was present in binding reaction b). oriC bound is defined as (cpm in the retarded band) × 100/(cpm in the retarded band + cpm in the unretarded oriC band).
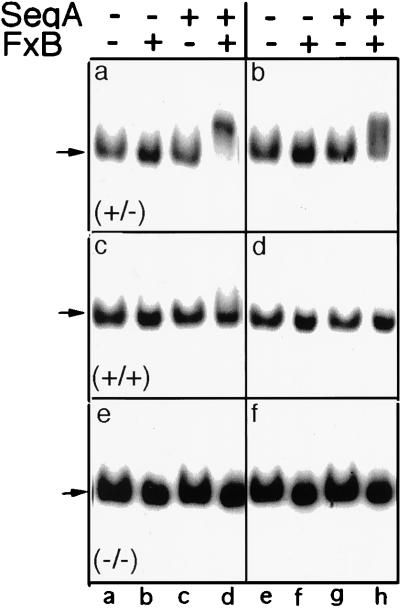
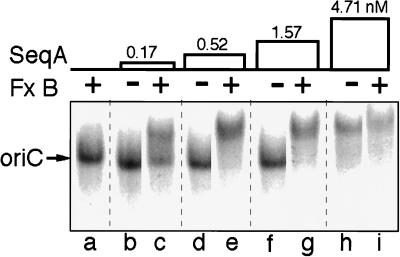
Figure 5.
Specificity of reconstituted system for hemimethylated oriC. Gel retardation assays were performed in the presence or absence of SeqA (2.9 nM) (a, c, and e) and/or fraction B (FxB, 40 ng/μl) (b, d, and f), as indicated. The concentrations of hemimethylated (a and b), fully methylated (c and d), or unmethylated (e and f) 32P-labeled oriC were 50 pM (lanes a–d) and 200 pM (lanes e–h). Autoradiograms were exposed for 12 hr (lanes a–d) or 3 hr (lanes e–h). Arrows indicate position of free oriC.
To further characterize the membrane component(s) responsible for oriC binding, we used several approaches to separate the membrane-associated binding activity from other membrane components. Greatest success was achieved with the use of chaotropes, agents that disrupt hydrophobic interactions (15, 16). Efficient release of the oriC-binding activity from the residual particulate membrane material was achieved by treatment of the membrane preparation with the moderate chaotropic agent KSCN at a concentration of 0.5 M (Fig. 1a, lane b). This solubilized 10–20% of the membrane protein and 0.4% of membrane lipid as determined by measurements on samples prepared from cells labeled with [2-3H]glycerol. Approximately 60% of the original membrane-associated oriC-binding activity was recovered in the KSCN extract, as shown by assays with limiting amounts of membrane and extract.
Dissociation of Binding Activity into Two Components.
The KSCN extract was further fractionated by exploiting its differential solubility characteristics. When KSCN was removed by dialysis, a precipitate formed that included approximately 50% of the total protein in the extract. The insoluble material recovered from the dialysis bag was resolubilized by treatment with KSCN and was used as fraction A in subsequent assays. The material that remained soluble after the dialysis procedure was called fraction B. The protein profiles of the two fractions were different, as shown by SDS gel electrophoretic analysis (data not shown).
Neither fraction A nor fraction B showed detectable oriC-binding activity when assayed at concentrations that were equivalent to the concentrations of the crude KSCN extract that showed strong binding activity in the same assay (Fig. 1b). However, when fraction A and fraction B were recombined prior to dilution into the binding mixture, oriC-binding activity was restored. Optimal binding occurred at a ratio of fraction A to fraction B of 1:2 (Fig. 1b, lanes e–g). Under these conditions, approximately 90% of the original activity of the KSCN extract was recovered in the reconstituted system (Fig. 1b, lane f).
Characterization of Fraction A and Fraction B.
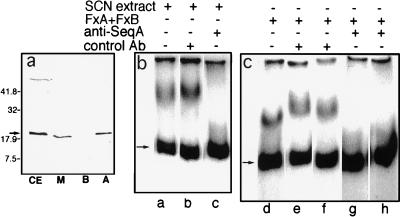
The following evidence indicated that SeqA was the active component of fraction A. (i) Immunoblot analysis with anti-SeqA antibody revealed that fraction A contained essentially all of the SeqA that was recovered from the original membrane preparation whereas fraction B contained no detectable SeqA (Fig. 2a). (ii) Anti-SeqA antibody inhibited oriC binding in the reconstituted system whereas control antiserum did not (Fig. 2 b and c). (iii) As further described below, purified SeqA was capable of substituting for fraction A in the reconstituted system.
Figure 2.
Effect of anti-SeqA antibody. (a) Anti-SeqA immunoblot analysis was performed on samples of cell extract (CE), membrane (M), fraction A (A), and fraction B (B). The amount of each sample applied to the gel corresponded to the amount of cell extract applied to the first lane to permit recoveries in each fraction to be estimated directly from the intensities of the SeqA bands. The arrow indicates the position of purified SeqA that was run on the same gel and stained with Coomassie brilliant blue. The band in the upper portion of the gel represents a cross-reacting component present in cell envelope preparations. The positions of molecular mass standards, expressed in kDa, are indicated on the left. (b and c) Gel retardation assays were performed on the KSCN extract (8 ng/μl, lanes a–c) or on a mixture of fraction A (Fx A, corresponding to 3 ng/μl of KSCN extract) and fraction B (Fx B, corresponding to 12 ng/μl of KSCN extract) (lanes d–h). Anti-SeqA antibody (lanes c, g, and h) or normal rabbit serum (lanes b, e, and f) was added to the sample to be assayed, in the assay buffer, and allowed to stand for 30 min at 4°C before being added to the assay mixture. Antisera were present at dilutions of 1:10 (lanes e and g) or 1:20 (lanes b, f, and h).
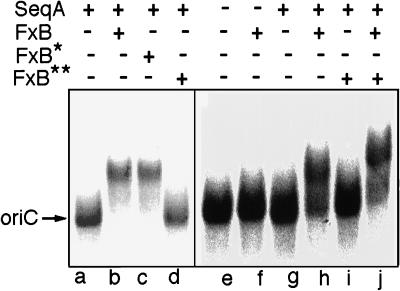
The active component of fraction B was nondialyzable, heat stable, and protease sensitive. When fraction B was preincubated with Pronase, oriC-binding activity was completely lost (Fig. 3, lanes d and i). When fraction B was treated identically but in the presence of a 10-fold lower concentration of Pronase (Fig. 3, lane c), there was no loss of activity. In a parallel experiment, fraction B that had not been protease treated was added to the reaction mixture after addition of the protease-treated fraction B (Fig. 3, lane j). This led to complete restoration of the binding activity, confirming that the apparent loss of activity of the protease-treated fraction B did not result from inactivation of SeqA by residual protease in the reaction mixture. These results indicate that the active component of fraction B is likely to be a protein. We tentatively name this factor SeqB. Fraction B had no detectable oriC-binding activity of its own when assayed at concentrations between 2.5 and 20 ng/μl in the absence of fraction A.
Figure 3.
Effect of Pronase on activity of fraction B. Gel retardation assays were performed in the presence or absence of SeqA (0.03 ng/μl) and/or fraction B (FxB, 10 ng/μl), as indicated. Pronase-treated fraction B (FxB* and FxB**) was prepared by incubating a mixture of fraction B (0.11 μg/μl) and Pronase in 10 mM Pipes (pH 6.5) for 30 min at room temperature followed by heating at 100°C for 6 min. In reaction c (FxB*), the concentration of Pronase was 0.009 μg/μl; in reactions d, i, and j (FxB**), Pronase concentration was 0.09 μg/μl. In reaction j, untreated fraction B was added to the reaction mixture after the addition of FxB**. Lanes a–d and e–j represent experiments done on separate days with different exposure times during autoradiography.
Direct evidence that SeqA was the active component of fraction A came from the finding that fraction A could be replaced by purified SeqA in the reconstituted binding system (Figs. 3 and 4). Binding of hemimethylated oriC at low SeqA concentrations showed an absolute dependence on the presence of SeqB over a 25–30-fold range of SeqA concentrations (Fig. 4). At high concentrations, SeqA was capable of binding hemimethylated oriC in the absence of added SeqB, as previously described (9). These and similar experiments established that SeqB increased the apparent affinity of hemimethylated oriC for SeqA by at least 30-fold.
Figure 4.
Effect of fraction B on oriC–SeqA interaction. Gel retardation assays were performed on the components indicated. Concentrations of SeqA (as nM) are indicated; fraction B (Fx B) was present, where indicated, at a concentration of 20 ng/μl.
SeqB-dependent binding to hemimethylated oriC was detectable as a shift of the 32P-labeled oriC band at oriC concentrations as low as 10 pM, the lowest concentration that was tested. In contrast, there was no detectable binding to fully methylated or unmethylated oriC at oriC concentrations up to 200 pM, the highest concentration that was tested (Fig. 5). Therefore, the affinity of the SeqA/SeqB system for fully methylated oriC is at least one to two orders of magnitude lower than for hemimethylated oriC.
At higher SeqA concentrations that do not require SeqB for binding to hemimethylated oriC, it has ben shown that SeqA can bind hemimethylated lacZ and, at even higher concentrations, SeqA can bind fully methylated oriC (9). The effect of SeqB under these conditions has not been examined.
Fraction B could not be replaced in the reconstituted system by 2–10 mM MgCl2 and the binding reaction was not affected by 2–10 mM EDTA, demonstrating that SeqB did not function by simply providing Mg2+ to the system.
DISCUSSION
We conclude from the present study that the high-affinity binding of hemimethylated oriC to E. coli membranes, which has previously been implicated in prevention of premature reinitiation at newly replicated chromosomal origins, requires both SeqA and a previously unrecognized factor, SeqB. The reconstituted SeqA/SeqB system retained the salient characteristics that distinguish the activity of the membrane-associated system. These include the ability to bind oriC at low SeqA concentrations and the high specificity for hemimethylated DNA. Taken together with the fact that most of the oriC-binding activity of the membrane preparation was recovered in the reconstituted SeqA/SeqB system, these observations provide the first evidence that the specific binding of hemimethylated oriC is likely to be mediated by a multiprotein complex that includes SeqA and SeqB or occurs in a localized membrane domain that includes these two components. The possibility that additional components may also be part of the high-affinity binding system cannot be excluded.
The results indicate that the role of SeqB is to activate the intrinsic oriC-binding activity of SeqA since SeqA at high concentrations was capable of binding hemimethylated oriC in the absence of SeqB. The requirement for SeqB could not be eliminated by increasing the concentration of hemimethylated oriC, suggesting that SeqB probably acts on SeqA rather than on the oriC substrate. Although it appears likely that the SeqA/SeqB system represents all or part of the machinery that normally sequesters hemimethylated oriC within the cell, proof of this will require identification and disruption of the seqB gene and characterization of the seqBnull phenotype.
The possible role of the membrane in the SeqA/SeqB-mediated sequestration process has not been fully characterized. The membrane association of SeqA and SeqB has thus far been defined primarily by their presence in a low-density crude membrane fraction. Although the membrane association of the hemimethylated oriC-binding activity of the crude membrane preparation had previously been rigorously established by sedimentation and flotation equilibrium analysis (6), a more rigorous characterization of the apparent membrane association of the SeqA/SeqB machinery will be needed to firmly establish the role of the membrane in the sequestration process.
Newly replicated chromosomal origins are refractory to the initiation of another round of replication until the appropriate time in the next cell cycle is reached. Support for the idea that this sequestration involves the association of hemimethylated oriC with membrane-associated SeqA includes the fact that sequestration of hemimethylated oriC in vivo and the ability of isolated E. coli membranes to bind hemimethylated oriC in vitro are both lost in ΔseqA cells (8, 9). However, it is striking that most of the cellular SeqA in broken cell preparations from growing cultures is recovered in the soluble fraction. In our hands, this accounts for approximately 70–90% of the total cellular SeqA, depending on growth conditions (unpublished data). The soluble material presumably represents cytosolic SeqA, although it could reflect membrane-associated SeqA that has been displaced from the membrane due to mechanical forces or to other factors in the preparative procedure.
The possibility that soluble and membrane-associated SeqA carry out different biological functions remains to be explored. Newly replicated oriC remains hemimethylated for about 30–40% of the cell cycle (4), and it seems clear that binding of hemimethylated oriC to SeqA plays a key role in its sequestration from premature reinitiation. On the other hand, initiation continues to be suppressed until the remaining 60–70% of the cycle has passed. The explanation for the continued eclipse period after oriC has become fully methylated is unknown. The continued failure of initiation could reflect unavailability of DnaA or other components required to trigger initiation (17) or could result from the association of fully methylated oriC with another oriC-binding protein. Alternatively, the continued sequestration of oriC from the initiation machinery could reflect a continued association of oriC with SeqA even after the new daughter strand is methylated, implying a possible role for cytosolic SeqA in this second-stage sequestration process.
In this regard it may be relevant that although purified SeqA has a strong preference for hemimethylated DNA, fully methylated oriC can bind to SeqA when the protein is present at higher concentrations (9). The binding to fully methylated oriC appears to be sequence specific since it was shown that SeqA fails to bind to fully methylated lacZ DNA. It has been suggested that the interaction of SeqA with fully methylated oriC may play a role in modulating initiation (9). If this is correct, it could explain the suppression of initiation that continues after oriC has been completely methylated.
Acknowledgments
We thank Jessica Epstein and Nancy Kleckner for the SeqA protein and Erik Boye for the anti-SeqA antibody. This work was supported by grants from the National Institutes of Health (GM53276) to L.R. and the Human Frontiers in Science Program (RG-386/95, to L.R. and M.K.).
ABBREVIATION
- KSCN
potassium thiocyanate
References
- 1.Crooke E. Cell. 1995;82:877–880. doi: 10.1016/0092-8674(95)90020-9. [DOI] [PubMed] [Google Scholar]
- 2.Skarstad K, Boye E. Biochim Biophys Acta. 1994;1217:111–130. doi: 10.1016/0167-4781(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 3.Ogden G, Pratt M, Schaechter M. Cell. 1988;54:127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- 4.Campbell J L, Kleckner N. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 5.Russell D, Zinder N. Cell. 1987;50:1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborti A, Gunji S, Shakibai N, Cubeddu J, Rothfield L. J Bacteriol. 1992;174:7202–7206. doi: 10.1128/jb.174.22.7202-7206.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landoulsi A, Malki A, Kern R, Kohiyama M, Hughes P. Cell. 1990;63:1053–1061. doi: 10.1016/0092-8674(90)90508-c. [DOI] [PubMed] [Google Scholar]
- 8.Lu M, Campbell J, Boye E, Kleckner N. Cell. 1994;77:1–20. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 9.Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 10.Boye E, Stokke T, Kleckner N, Skarstad K. Proc Natl Acad Sci USA. 1996;93:12206–12211. doi: 10.1073/pnas.93.22.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrick J, Kern R, Guha S, Landoulsi A, Fayet O, Malki A, Kohiyama M. EMBO J. 1994;13:4695–4703. doi: 10.1002/j.1460-2075.1994.tb06793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.de Boer P A J, Crossley R E, Rothfield L I. Proc Natl Acad Sci USA. 1990;87:1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brendler T, Abeles A, Austin S. EMBO J. 1995;14:4083–4089. doi: 10.1002/j.1460-2075.1995.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moldow C, Robertson J, Rothfield L I. J Membr Biol. 1972;10:137–152. doi: 10.1007/BF01867850. [DOI] [PubMed] [Google Scholar]
- 16.Karnauchov I, Herrmann R G, Klosgen R B. FEBS Lett. 1997;408:206–210. doi: 10.1016/s0014-5793(97)00427-4. [DOI] [PubMed] [Google Scholar]
- 17.Lobner-Olesen A, Hansen F G, Rasmussen K V, Martin B, Kuempel P L. EMBO J. 1994;13:1856–1862. doi: 10.1002/j.1460-2075.1994.tb06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]