Abstract
A sensitive in vitro crosslinking technique using a photoactive derivative of the chimeric activator LexA-E2F-1 was used to identify yeast proteins that might influence the response of RNA polymerase II to transcriptional activators. We found that a novel yeast protein, Xtc1p, could be covalently crosslinked to the activation domain of LexA-E2F-1 when this derivatized activator was bound to DNA upstream of an activator-responsive RNA polymerase II promoter. Because affinity chromatography experiments showed that Xtc1p also bound directly and specifically to the activation domains of E2F-1, the viral activator VP16, and the yeast activator Gal4p and copurified with the RNA polymerase II holoenzyme complex, Xtc1p may modulate the response of RNA polymerase II to multiple activators. Consistent with this notion, yeast strains deleted for the XTC1 gene exhibited pleiotropic growth defects, including temperature sensitivity, galactose auxotrophy, and a heightened sensitivity to activator overexpression, as well as an altered response to transcriptional activators in vivo.
RNA polymerase II (RNA pol II), the enzyme responsible for synthesis of messenger RNA in eukaryotes, is controlled through the action of positive- and negative-acting regulatory proteins (1, 2). Of primary importance are the large number of sequence-dependent DNA-binding proteins that regulate the activity of RNA pol II in a gene-specific manner. For positive-acting gene regulators, a defined region of the protein, its activation domain, often retains the ability to stimulate transcription when fused to a heterologous DNA-binding domain such as that of the yeast activator Gal4p (3) or the bacterial repressor LexA (4). Activation domains frequently exhibit a characteristic amino acid composition. For example, the activation domains of many mammalian, viral, and yeast activators are rich in acidic amino acids (3, 5, 6), although negative charge is neither sufficient nor essential for activation domain function (7, 8). Because activation domains often retain the ability to potentiate transcription in evolutionarily divergent species, the mechanism(s) that mediate transcriptional activation may be conserved.
Studies on the control of transcription in the yeast Saccharomyces cerevisiae have led to the suggestion that one key function of activators is to facilitate the recruitment of a preassembled RNA pol II holoenzyme complex to promoter DNA (9–12). Indeed, artificial tethering of individual components of the RNA pol II transcriptional machinery to promoter DNA as a result of fusion to a DNA-binding domain can result in an activated level of transcription in the absence of a bona fide activation domain (11–14). Activators also appear to stimulate the activity of RNA pol II subsequent to its engagement with the promoter, either by enhancing the rate of transcript initiation, promoter clearance, and/or chain elongation (15). It is generally assumed that this potentiation of transcription arises, at least in part, though direct contact of activators with components of the RNA pol II transcriptional machinery. Putative targets of activators in both yeast and higher eukaryotes include a number of the general transcription factors (16–19) that mediate the interaction of RNA pol II with the core promoter DNA sequences as well as a range of coactivator proteins now known to have either intrinsic or associated chromatin remodeling activities (for example, refs. 20 and 21).
Activators, in turn, can be regulated through direct association with specific repressor proteins. For example, the yeast activator Gal4p is maintained in an inactive state in the absence of galactose by the direct and specific interaction of the inhibitory protein Gal80p with its C-terminal activation domain (22). A similar form of control may operate in higher eukaryotes as well. Both the human tumor suppressor protein p53 and the cell cycle-regulated activator E2F-1 are negatively regulated through interaction of their activation domains with inhibitory proteins Mdm-2 and Rb, respectively (23, 24).
To identify additional proteins that might influence the response of RNA pol II to transcriptional activators, we developed a photochemical crosslinking technique to detect proteins in close proximity to the derivatized activation domain of the sequence-specific transactivator LexA-E2F-1 in an in vitro transcription system. Using this approach, we have identified a novel yeast protein Xtc1p that appears to act as a negative regulator of transcriptional activation in vivo.
MATERIALS AND METHODS
Yeast Cell Extracts.
For our initial crosslinking experiments, whole-cell extracts were prepared from the haploid yeast strain DPY213 (a leu2 ura3 his3 trp1 lys2 taf130Δ∷TRP1Δ + pRS313-flu3-TAF130) (25) and fractionated by chromatography on a Bio-Rex 70 ion-exchange column as described for the purification of yeast RNA pol II holoenzyme (26). The transcription-competent 0.6-M potassium acetate fraction was used in the crosslinking experiments after dialysis against transcription buffer (50 mM Hepes⋅KOH/90 mM potassium acetate/10% glycerol/10 mM Mg acetate/2 mM EGTA/2 mM DTT, pH 7.6) and had a final protein concentration of ≈20 mg/ml. Similar column fractions were prepared from isogenic wild-type and xtc1Δ strains in which the XTC1 ORF had been replaced with either the LEU2 or TRP1 gene by standard gene-replacement procedures. Affinity-purified yeast RNA pol II holoenzyme was a generous gift from G. Pan and J. Greenblatt (27).
Preparation of Photoreactive LexA-E2F-1.
The LexA-E2F-1 fusion protein was expressed in Escherichia coli cells by using a modified pET-19b expression vector (Novagen) encoding tandem N-terminal poly(10)His and heart muscle kinase recognition tags and was purified to homogeneity by metal chelate chromatography (28). The recombinant protein was labeled with [γ-32P]ATP (6,000 Ci/mmol; New England Nuclear) to a specific activity of 2 × 105 cpm/pmol by using heart muscle kinase (Sigma) (28). The labeled protein was bound to Ni2+-NTA agarose beads (Qiagen) and washed extensively with buffer B (20 mM Hepes⋅NaOH, pH 7.9/100 mM NaCl/20% glycerol/0.2 mM EDTA). A 5-fold molar excess (relative to protein) of maleimide-4-benzophenone (MBP) (Sigma) dissolved in dimethyl formamide then was added under reduced lighting conditions. After incubation in the dark for 2 h at 25°C, the beads were washed with buffer B and eluted with buffer containing 1 mM DTT and 0.5 M imidazole. Derivatization was determined to be 90% (28).
Crosslinking.
Photoreactive LexA-E2F-1 (3 pmol; ≈5 × 105 cpm) was added under reduced lighting to microtubes containing 10 μl of yeast transcription extract (0.2 mg protein)/0.5 pmol of template DNA/3 pmol of recombinant yeast TFIIA (28), in a final volume of 25 μl. The mixture(s) were placed 5 cm under an inverted UV-transilluminator (Fotodyne model 3–3100, Hartland, WI) and UV-irradiated for 12 min at 25°C. The reaction products were fractionated on 7.5% polyacrylamide gels containing SDS. The gels were dried and exposed to film with an amplifying screen for 12 h at 25°C.
Affinity Chromatography, Immunoblotting, and Immunoprecipitation.
Protein ligands were coupled to AffiGel 10 resin (Bio-Rad) to a final concentration of 2 mg/ml. Microcolumns were prepared as described (29) and loaded with either 0.4 mg of yeast transcription extract or 2 μg of purified recombinant Xtc1p. The columns were washed with 10 column volumes of transcription buffer and eluted with 3 column volumes of buffer containing 1 M NaCl. To prepare protein for microsequencing, 40 ml of Bio-Rex 70-fractionated yeast extract was loaded on a 2-ml LexA-E2F-1 affinity column. The bound yeast proteins were eluted by 1 M NaCl, concentrated by TCA precipitation, and fractionated on a 12.5% polyacrylamide gel. Protein sequencing was as described (30). Xtc1p was detected on immunoblots by enhanced chemiluminescence (ECL; Amersham) using polyclonal serum from rabbits immunized with recombinant Xtc1p. To immunoprecipitate Xtc1p, 20 μl of a standard crosslinking reaction was diluted with 500 μl of TTBS (0.05% Tween 20/10 mM Tris⋅HCl, pH 7.9/0.5 M NaCl) and incubated with 2 μl of rabbit antisera for 4 h on ice and then with protein A beads. The beads were boiled in SDS gel sample buffer before electrophoretic separation of bound proteins.
Yeast Cell Manipulations.
Yeast cells were transformed by using the Li acetate technique (31). The XTC1 ORF was replaced with a LEU2 or a TRP1 gene cassette in the diploid W3031 strain LP112 (a/α can1–100/can1–100 his3–11, 15/his3–11, 15 leu2–3, 112/leu2–3, 112 trp1–1/trp1–1 ura3–1/ura3–1 ade2–1/ade2–1) or a haploid derivative (32). β-Galactosidase assays (32) were normalized to the OD595 of the cultures and the assay time. A yeast expression vector for Gal4-E2F-1 under control of the ADH1 promoter was kindly provided by A. Pearson and J. Greenblatt (33).
RESULTS
Crosslinking of LexA-E2F-1 to Yeast Proteins in a Cell-Free Transcription System.
To better characterize the range of protein–protein interactions that influence the response of RNA pol II to sequence-specific activators, we sought to identify components of the yeast RNA pol II transcriptional machinery that can interact with, or are positioned nearby, the activation domain of a promoter-bound activator during the process of transcriptional activation in vitro. Our approach involved the selective placement of a photoreactive crosslinking side chain on the single cysteine residue within the activation domain of the chimeric activator LexA-E2F-1 (28), an activator that consists of the acidic C-terminal activation domain (amino acid residues 400–437) of the human activator E2F-1 (24) fused to the bacterial DNA-binding protein LexA. This chimeric protein is a potent sequence-specific activator of transcription by RNA pol II in vitro (ref. 28; see below) and in vivo (Fig. 5).
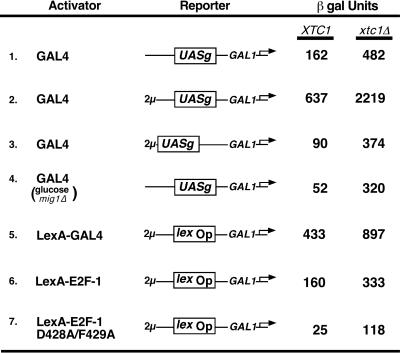
Figure 5.
Enhanced activation of transcription in Xtc1p-deficient yeast. β-Galactosidase activities in permeabilized yeast cells containing the following lacZ fusion reporter genes: lines 1 and 4, a single copy of the integrated plasmid RY171 (39), which contains Gal4-binding sites derived from the GAL1–10 UASg element upstream of the GAL1 core promoter; line 2, a 2-μ derivative of RY171; line 3, the 2-μ plasmid pJK101 (4), a derivative of RY171 in which the UASg is located a further 100 nt distal to the GAL1 promoter; and lines 5–7, the 2-μ plasmid p18–40 (4), which has a single LexA operator sequence upstream of the GAL1 core promoter. LexA-E2F-1 and LexA-GAL4 were expressed from the ADH1 promoter on 2-μ vectors (4). The strain used in line 4 had the MIG1 gene deleted. Cells were grown in the presence of 2% (wt/vol) galactose supplemented with either 2% (wt/vol) sucrose (lines 1–3, 5–7) or glucose (line 4) and were harvested at mid-log phase. Activities are expressed in Miller units (32); SD values were less than 20%.
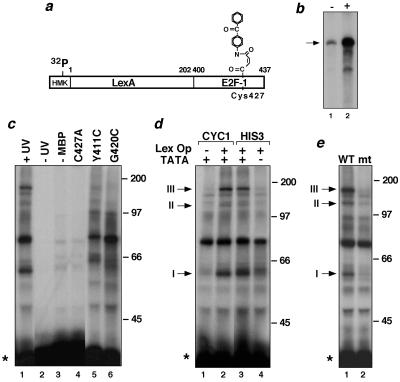
Purified recombinant LexA-E2F-1 was labeled to a high specific activity with [32P]ATP. Next, the cysteine residue located within the E2F-1 activation domain (residue 427 in E2F-1; refs. 24 and 28) was selectively derivatized with the hetero-bifunctional, photoreactive chemical crosslinking reagent MBP (Fig. 1a) (28). This derivatization of the activator did not affect its ability to potentiate transcription by RNA pol II in a yeast in vitro transcription system (Fig. 1b; see also ref. 28).
Figure 1.
Selective crosslinking of a transcriptional activator to yeast proteins in an in vitro transcription system. (a) Schematic of the 32P-labeled, MBP-derivatized, photoreactive LexA-E2F-1 activator protein. (b) Autoradiogram of RNA transcripts produced by a yeast transcription extract with (+) and without (−) photoreactive LexA-E2F-1 added. The DNA template had two LexA-binding sites upstream of the CYC1 promoter (28). (c) Autoradiogram of SDS-gel-fractionated crosslinking reactions containing photoreactive LexA-E2F-1, yeast transcription extract, and the DNA template used in b. All reactions, except for lane 2, were subject to UV irradiation. In lane 3, the activator had not been treated with MBP. Crosslinking by LexA-E2F-1 derivatives without a Cys residue or mutated to have a Cys at amino acid position 411 or 420 in E2F-1 also are shown (lanes 4–6). Mr of protein markers are given in kDa; LexA-E2F-1 is indicated with a star. (d) Promoter-dependent crosslinking by LexA-E2F-1. Crosslinking by photoreactive LexA-E2F-1 in the presence of DNA templates with (+) or without (−) LexA-binding sites or a TATA-box initiator element. The templates had either the CYC1 (lanes 1 and 2) or the HIS3 TR (lanes 3 and 4) core promoter elements (28). Promoter-dependent, activator-crosslinked complexes I, II, and III are indicated. (e) Impaired crosslinking by the D428A/F429A E2F-1 activation domain mutant (lane 2) relative to the wild-type activator (lane 1).
To induce crosslinking between LexA-E2F-1 and yeast proteins in close proximity to the E2F-1 activation domain during transcriptional activation, photoreactive LexA-E2F-1 was incubated with a partially purified preparation of the yeast RNA pol II holoenzyme complex and a DNA template bearing LexA-binding sites upstream of the CYC1 core promoter and irradiated briefly with UV light. As seen in Fig. 1c, several labeled complexes with distinct electrophoretic mobilities, presumably consisting of LexA-E2F-1 covalently bound to different yeast proteins, could be resolved by SDS/PAGE (lane 1). Control reactions confirmed that formation of these activator-crosslinked complexes depended on irradiation of the extract with UV light (Fig. 1c, lane 2) and on pretreatment of the activator with the crosslinker (lane 3). Substitution of the reactive cysteine residue in LexA-E2F-1 with alanine also abrogated crosslinking of LexA-E2F-1 to these yeast proteins (lane 4), indicating that the crosslinking was mediated solely through the derivatized cysteine residue located within the E2F-1 activation domain. Furthermore, the crosslinking exhibited by LexA-E2F-1 appeared to be selective since the pattern of crosslinked complexes was altered by repositioning the reactive cysteine residue and, therefore, the crosslinker, at residues 411 or 420 within the E2F-1 activation domain (lanes 5 and 6)—changes that did not substantially affect the ability of the activator to stimulate transcription (23, 28).
To establish whether the crosslinked complexes resulted from an interaction between DNA-bound LexA-E2F-1 and yeast proteins present at the adjacent RNA pol II promoter, the same crosslinking reaction was performed by using DNA templates that lacked either LexA-binding sites or a TATA-box promoter element. The results shown in Fig. 1d indicate that three of the more prominent activator-crosslinked complexes (I, II, and III) formed preferentially when the DNA template contained LexA sites and the TATA box. The appearance of these complexes was reduced markedly using templates lacking either of these elements (compare lanes 1 and 4 with lanes 2 and 3).
The promoter-dependent crosslinking exhibited by photoreactive LexA-E2F-1 suggested that one or more of the crosslinked yeast proteins might be associated with the response of RNA pol II to this activator. Therefore, the effects of introducing a double point mutation (D428A/F429A) into the E2F-1 activation domain, a change that impairs the ability of LexA-E2F-1 to activate transcription in vitro and in vivo (ref. 28; Fig. 5), was analyzed. As seen in Fig. 1e, this mutant activator was impaired in its ability to form complexes I, II, and III. It would appear, therefore, that the promoter-dependent crosslinking exhibited by LexA-E2F-1 may reflect important interactions mediated by this activator with the RNA pol II transcription machinery.
Identification of Xtc1p as an Activation-Domain Binding Protein.
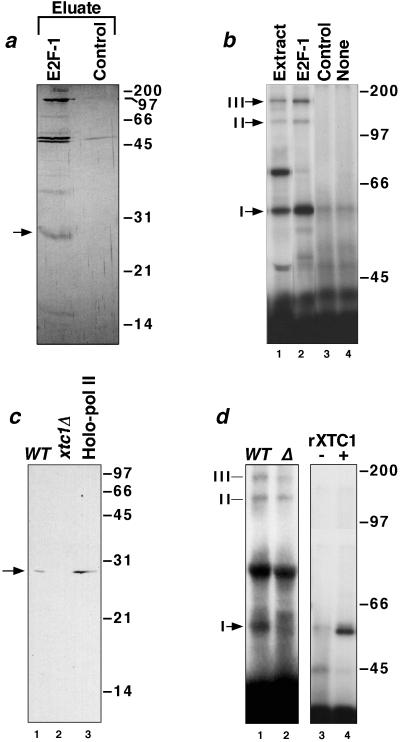
To identify yeast proteins that crosslinked to LexA-E2F-1, we used affinity chromatography as a purification step. Yeast cell extract, enriched for components of the RNA pol II holoenzyme complex by chromatography on Bio-Rex 70 (26), was fractionated further on an affinity column matrix containing immobilized LexA-E2F-1 as ligand. After extensive washing of the matrix, activator-bound proteins were eluted with a high-salt buffer and visualized by silver staining (Fig. 2a). Several proteins were found to be retained selectively on the LexA-E2F-1 affinity column (compare lane 1 with the eluate of the LexA control column, lane 2). UV irradiation of this eluate in the presence of photoreactive LexA-E2F-1 resulted in the formation of three crosslinked complexes (Fig. 2b, lane 2) with the same mobilities as that of complexes I, II, and III formed with the input yeast extract (lane 1), suggesting that the crosslinkable yeast proteins were retained specifically by the affinity matrix. In contrast, none of these complexes was observed after a similar crosslinking with the high-salt eluate from the control LexA affinity column (lane 3) or in the absence of added yeast extract (lane 4).
Figure 2.
Purification and cloning of Xtc1p. (a) Silver-stained SDS gel of the high-salt eluates from a LexA-E2F-1 (lane 1) and a control LexA (lane 2) affinity column. A 28-kDa yeast polypeptide that binds specifically to the E2F-1 activation domain is indicated. (b) Crosslinking by photoreactive LexA-E2F-1 in a yeast transcription extract (lane 1), in eluates from the LexA-E2F-1 and LexA affinity columns (lanes 2 and 3), or in the absence of added protein (lane 4). (c) Immunoblots with anti-Xtc1p of extracts (50 μg) prepared from isogenic wild-type (lane 1) and xtc1Δ (lane 2) yeast strains and of an aliquot (100 μg) of TFIIS affinity-purified yeast RNA pol II holoenzyme (lane 3) (27). (d) SDS-gel analysis of crosslinking by photoreactive LexA-E2F-1 to proteins in extracts from wild-type and xtc1Δ yeast cells (lanes 1 and 2) and crosslinking by photoreactive LexA-E2F-1 in the absence and presence of 200 ng of purified recombinant Xtc1p (lanes 3 and 4).
Because it seemed that the 28-kDa protein present in the LexA-E2F-1 affinity column eluate (Fig. 2a) might account for the formation of complex I in the crosslinking reactions with photoreactive LexA-E2F-1, we scaled up the affinity-purification procedure to obtain a sufficient amount of this protein for direct identification by peptide microsequencing. The 28-kDa band was excised from a preparative SDS gel and a 17-mer peptide sequence, LIQRVGNIAREESVILK, of high confidence obtained (see Materials and Methods). This sequence was found to match perfectly to a portion of an ORF, YDR296W, located on chromosome IV of S. cerevisiae that encodes a previously uncharacterized gene product of 226 aa (Mr = 26,906, pI = 9.64). Although the predicted amino acid sequence of the putative full-length gene product, which we have named Xtc1 for crosslinked transcription component 1, did not provide additional insight as to its cellular function, it did exhibit limited sequence similarity (20–25% identity over residues 75–216 in Xtc1p) to both the yeast (34) and human (GenBank accession no. X97795) DNA repair-associated helicase Rad54 in a region encompassing Rad54’s canonical helicase motifs IV, V, and VI. The functional significance of this sequence similarity remains unclear because the much smaller Xtc1p protein lacks motifs I, Ia, II, and III, which are required for helicase/ATPase activity. As indicated above, Xtc1p copurified with yeast RNA pol II and other components of the RNA pol II holoenzyme complex upon fractionation of a crude yeast cell lysate on Bio-Rex 70 resin. Subsequently, we found that Xtc1p also was present in a more highly purified preparation of yeast RNA pol II holoenzyme isolated by affinity chromatography on immobilized yeast TFIIS (Fig. 2c). Xtc1p therefore may be a component of the RNA pol II transcriptional machinery targeted by transcriptional activators.
To confirm that the activator-crosslinked complex I formed in the in vitro transcription system did indeed consist of LexA-E2F-1 covalently linked to Xtc1p, the following experiments were performed. We first established that a transcription extract prepared from yeast cells deleted for the XTC1 gene failed to generate complex I in crosslinking experiments with photoreactive LexA-E2F-1 (Fig. 2d, compare lanes 1 and 2), although formation of the other activator-crosslinked complexes was largely unaffected. We also showed that recombinant Xtc1 protein, expressed in and purified from E. coli cells, was by itself capable of forming a crosslinked complex with photoreactive LexA-E2F-1 (lane 4) with the same electrophoretic mobility on an SDS-polyacrylamide gel as that of complex I. In addition, we found that crosslinked complex I formed with a wild-type yeast transcription extract could be immunoprecipitated selectively with antibodies raised against recombinant Xtc1p but not with preimmune serum (data not shown).
Activator-Binding Activities of Xtc1p.
To further assess the activation domain-binding activities of Xtc1p, protein affinity chromatography was used. As anticipated from our crosslinking and purification experiments (Figs. 1 and 2), immobilized LexA-E2F-1 was found to bind strongly to and retain Xtc1p in a yeast cell extract (Fig. 3 Upper). Since LexA-E2F-1 also bound well to purified recombinant Xtc1p (Lower), this interaction appears to be direct. In contrast, both the cellular and recombinant forms of Xtc1p bound poorly to the D428A/F429A mutant derivative of LexA-E2F-1 (lane 4) that is impaired for both transcriptional activation in vivo (Fig. 5, line 7) (24) and the ability to form crosslinked complex I in vitro (Fig. 1e).
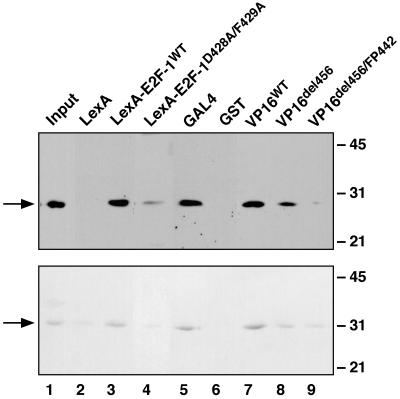
Figure 3.
Xtc1p interacts directly with the activation domains of several different activators. (Upper) Immunoblot analysis of Xtc1p (arrow) in the input yeast extract (lane 1) and high-salt eluates (lanes 2–9) from a series of affinity columns. The column ligand is indicated above each lane. (Lower) Coomassie blue-stained SDS gel showing the input recombinant Xtc1 protein (lane 1) and high-salt eluates (lanes 2–9) from a duplicate set of affinity columns. For both panels, 20% of the input sample and 50% of the column eluate were analyzed.
To establish whether Xtc1p could interact with the activation domains of other transcriptional activators, additional affinity chromatography experiments were performed using the activation domains of the yeast activator Gal4p (amino acids 841–874) (6) and the herpes viral protein VP16 (amino acids 413–490) (3) as ligands. As with immobilized LexA-E2F-1, both the native and recombinant forms of Xtc1p were retained efficiently on affinity columns bearing either of these two other activation domains (Fig. 4, lanes 5 and 7). Furthermore, similar to the effects of the D428A/F429A mutation on binding by the E2F-1 activation domain, mutation of the Phe-442 residue in the VP16 activation domain to proline or truncation at amino acid 456 greatly reduced the affinity of VP16 for Xtc1p (lanes 8 and 9). These alterations have been shown previously to impair the ability of VP16 to activate transcription (7). Thus, the ability of both VP16 and E2F-1 to interact with Xtc1p correlates positively with their ability to stimulate transcription.
Figure 4.
Xtc1p is required for normal cell growth. (a) Growth of wild-type and xtc1Δ yeast strains on glucose at 30°C and 37°C. (b) Impaired growth of xtc1Δ yeast cells on galactose and restoration of growth by ectopic expression of Xtc1p. Cells were transformed with either a control vector (pADH1) or this vector expressing Xtc1p under control of the ADH1 promoter (pADH1-XTC1). (c) Impaired growth of xtc1Δ cells after overexpression of the strong activator Gal4-E2F-1 (33) but not the DNA-binding domain of Gal4p alone (GAL4-DBD).
Growth and Transcription Defects in Yeast Strains Deleted for the XTC1 Gene.
To better assess the physiological function of Xtc1p, a targeted deletion of the XTC1 gene was performed. As observed with yeast strains deleted for other accessory components of the RNA pol II transcriptional machinery, such as the Srb, Ada, and Swi/Snf classes of transcription cofactors (21, 35, 36), yeast strains deleted for the XTC1 gene (xtc1Δ) exhibited a variety of growth defects. For example, the xtc1Δ cells grew more slowly than wild-type cells on rich media and this growth was temperature-sensitive (Fig. 4a). The xtc1Δ cells also were unable to grow on media containing galactose as the carbon source, although viability was restored by plasmid-based expression of Xtc1p (Fig. 4b).
To determine whether the growth defects exhibited by the Xtc1p-deficient yeast cells were associated with an impaired response of RNA pol II to transcriptional activators, we assessed the ability of isogenic wild-type and xtc1Δ strains to support activated levels of transcription in vivo. Because the activation domain of Gal4p, like that of E2F-1, bound to Xtc1p in our affinity chromatography experiments, we first evaluated the ability of endogenous Gal4p to activate transcription in wild-type and xtc1Δ yeast cells. Surprisingly, Gal4p was found to activate transcription of three different Gal4p-responsive lacZ reporter genes three to five times more efficiently in the xtc1Δ cells than it did in wild-type cells (Fig. 5, lines 1–3). This increased responsiveness of RNA pol II to Gal4p in the absence of Xtc1p appears to be because of a general enhancement of activation domain potential in the mutant cells since a LexA protein fusion bearing the C-terminal activation domain of Gal4p also stimulated transcription more effectively in a xtc1Δ mutant background (Fig. 5, line 5). LexA-E2F-1 and the D428A/F429A mutant form of LexA-E2F-1 also were found to activate transcription more effectively in the xtc1Δ cells (lines 6 and 7). Like extracts prepared from Xtc1p-containing cells (Fig. 1b), extracts from xtc1Δ cells also showed enhanced transcription in vitro in response to addition of the activator LexA-E2F-1 (data not shown).
Deletion of the XTC1 gene also had a pronounced effect in relieving glucose-mediated repression of transcriptional activation by Gal4p in cells that also lacked the MIG1 gene product (line 4), similar to effects observed after deletion of the SRB8, SRB10, and SRB11 genes (37), which encode regulatory subunits of the RNA pol II holoenzyme complex. However, as there was no measurable promoter activity from the Gal4p-responsive reporter genes in a gal4Δ xtc1Δ strain (data not shown), this increase in transactivation potential in the absence of Xtc1p was not a consequence of constitutive transcription. Together, the results of this in vivo transcription analysis suggest that Xtc1p functions as a repressor of transcriptional activation.
As overexpression of strong transcriptional activators had been shown previously to inhibit RNA pol II-dependent transcription and to impair yeast cell growth, a phenomenon referred to as “squelching” (38), we reasoned that the pleiotropic defects exhibited by xtc1Δ strains might have resulted, at least in part, from the hyperactivity of one or more cellular activators in the absence of the inhibitory function of Xtc1p. Consistent with this notion, we found that the growth of xtc1Δ cells was impaired dramatically relative to that of wild-type cells when the potent chimeric activator Gal4-E2F-1 (amino acids 387–437 of E2F-1 fused to the DNA-binding domain of Gal4p) was expressed under control of the ADH1 promoter (Fig. 4c).
DISCUSSION
One current view of the process of activating transcription by RNA pol II is that promoter-bound activator proteins stimulate transcription by interacting directly with components of the RNA pol II transcriptional machinery (9–11). These interactions may hasten the assembly and/or alter the activity of preinitiation complexes at nearby promoters. In addition, several large, multiprotein complexes, such as those containing the Sw1/Snf (40) or Ada (21) proteins, some of which may be associated with RNA pol II in vivo (41), are thought to contribute to efficient activation of many yeast genes by virtue of their ability to remodel chromatin structure at promoters.
Although there is already documentation of a number of interactions of activators with different components of RNA pol II machinery, we reasoned that the crosslinking approach used in this study might be a useful method to reveal new aspects of the activation process. We anticipated that the identification of proteins that are capable of promoter-dependent crosslinking to a DNA-bound transcriptional activator might either confirm the involvement of previously described interactions in the activation process (16–19) or identify new proteins with undiscovered roles in transcription. In this report, we have identified one of several yeast proteins that can be crosslinked to a promoter-bound form of the chimeric activator LexA-E2F1. This crosslinked protein, Xtc1p, is capable of interacting tightly and specifically with the activation domains of a number of different activators and appears to play a physiologically significant role, negatively regulating the potency of activators.
Our previous studies had shown that a photoreactive derivative of LexA-E2F1 could be crosslinked to the TATA-box-binding protein TBP when both LexA-E2F1 and TBP were bound to adjacent sites on promoter DNA (28). These experiments, and others using affinity chromatography, had indicated that TBP and the general transcription factor TFIIH might each be contacted directly by this activator (33). Therefore, we had anticipated that when the crosslinking experiments were performed in the context of a complete activator-responsive system, some or all of these same interactions with components of the RNA pol II holoenzyme might be detected. Although the first of these crosslinked proteins identified, Xtc1p, is similar in size to yeast TBP polypeptide, there was no evidence for an appropriately sized crosslinked complex containing TBP, even in the xtc1Δ cell extract. The lack of crosslinking to TBP in these current experiments using a complete transcription system may have resulted from preferential detection in our experiments of interactions that occur after the initial formation of a RNA pol II preinitiation complex at the promoter. Alternatively, the addition of TFIIA to the extracts in the crosslinking experiments may have inhibited formation of this particular crosslink as it did in our previous studies (28). Furthermore, derivatization of the Cys-427 position may not be optimal for crosslinking to TBP in the context of the complete pol II holoenzyme. Other differently sized proteins appeared to be crosslinked when the Y411C and G420C derivatives were used (Fig. 1c).
The crosslinking experiments reported here certainly do not rule out the importance of direct activator contacts with the general transcription factors. Nevertheless, several lines of evidence suggest that Xtc1p also mediates some important aspect of transcriptional control. First, not only did Xtc1p interact with the activation domain of E2F-1 in a promoter-dependent manner, but it also bound well to the activation domains of Gal4p and VP16 in affinity chromatography experiments. That these latter interactions were observed in the absence of promoter DNA is presumably a result of the significantly higher activator concentrations (≈100 μM) used in the affinity chromatography experiments compared with those in crosslinking reactions (≈100 nM), where promoter dependence was observed. Second, the ability of Xtc1p to interact with these activators correlated well with the ability of each activator to stimulate transcription. Third, Xtc1p copurified with the cellular RNA pol II holoenzyme complex through steps of Bio-Rex 70, DEAE-Sepharose, and TFIIS affinity chromatography. Finally, both the increased transcriptional response with a number of different reporter gene constructs and the growth defects exhibited by xtc1Δ strains are consistent with the loss of an inhibitor of transactivation. Taken together, these data implicate Xtc1p as a negative regulator of the transcriptional activation process. Other components of the RNA pol II holoenzyme complex, namely, Sin4p, Rox3p, Srb8p, and Srb9p, also have been implicated in the repression of transcription (42).
The mechanism by which Xtc1p acts to down-regulate the response of yeast RNA pol II to activators presently is unclear. One possibility is that Xtc1p functions as a direct inhibitor of activation by competing with general transcription factors and coactivators for binding to activation domains. Alternatively, as with several other components of the yeast RNA polymerase II holoenzyme, Xtc1p may act as a more general repressor of transcription whose inhibitory effects are relieved by contact with activators. A third possibility is that Xtc1p inhibits transcriptional activation indirectly by influencing the stability of the activator to which it binds, just as the mammalian Mdm2 protein modifies the stability of p53 (43, 44). Our current efforts, which are focused on elucidating the mechanism by which Xtc1p represses transcription, as well as on the identification of the other yeast proteins that crosslinked to LexA-E2F1, should lead to a more complete understanding of the regulation of transcription by RNA polymerase II.
Acknowledgments
We thank R. Brent, H. Ronne, M. Johnston, J. Archambault, and A. Pearson for generously providing plasmids, C. Kelley for technical assistance with protein microsequencing, D. Jansma for help in tetrad analysis, and B. Andrews, J. Archambault, and J. Greenblatt for helpful discussion. This work was supported by a grant to C.J.I. from the Medical Research Council of Canada. R.K. was supported by National Institutes of Health Grant CA45508. A.E. was the recipient of a Medical Research Council of Canada Studentship.
ABBREVIATIONS
- RNA pol II
RNA polymerase II
- MBP
maleimide-4-benzophenone
References
- 1.Struhl K. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 2.Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 3.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature (London) 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 4.Brent R, Ptashne M. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 5.Hope I A, Struhl K. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 6.Ma J, Ptashne M. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 7.Cress W D, Triezenberg S J. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 8.Leuther K K, Salmeron J M, Johnston S A. Cell. 1993;72:575–585. doi: 10.1016/0092-8674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- 9.Greenblatt J. Curr Opin Cell Biol. 1997;9:310–319. doi: 10.1016/s0955-0674(97)80002-6. [DOI] [PubMed] [Google Scholar]
- 10.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 11.Barberis A, Pearlberg J, Simkovitch N, Farrell S, Reinagl P, Barnard C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee S, Struhl K. Nature (London) 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 13.Klages N, Strubin M. Nature (London) 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 14.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 15.Bentley D L. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 16.Stringer K F, Ingles C J, Greenblatt J. Nature (London) 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 17.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 18.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Mol Cell Biol. 1994;10:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 20.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 21.Roberts S M, Winston F. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J, Ptashne M. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 23.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 24.Flemington E K, Speck S H, Kaelin W G. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poon D, Campbell A M, Bai Y, Weil P A. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 26.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 27.Pan G, Aso T, Greenblatt J. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 28.Emili A, Ingles C J. J Biol Chem. 1995;270:13674–13680. doi: 10.1074/jbc.270.23.13674. [DOI] [PubMed] [Google Scholar]
- 29.Emili A, Greenblatt J, Ingles C J. Mol Biol Cell. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, Kobayashi R, Bishop J M. Proc Natl Acad Sci USA. 1996;93:8425–8430. doi: 10.1073/pnas.93.16.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allison L A, Ingles C J. Proc Natl Acad Sci USA. 1989;86:2794–2798. doi: 10.1073/pnas.86.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson A, Greenblatt J. Oncogene. 1997;15:2643–2658. doi: 10.1038/sj.onc.1201451. [DOI] [PubMed] [Google Scholar]
- 34.Emery H S, Schild D, Kellogg D E, Mortimer R K. Gene. 1991;104:103–106. doi: 10.1016/0378-1119(91)90473-o. [DOI] [PubMed] [Google Scholar]
- 35.Carlson M. Annu Rev Cell Dev. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Peterson C L, Herskowitz I. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 37.Balciunas D, Ronne R. Nucleic Acids Res. 1995;23:4421–4425. doi: 10.1093/nar/23.21.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gill G, Ptashne M. Nature (London) 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 39.Yocum R R, Hanley S, West R, Jr, Ptashne M. Mol Cell Biol. 1984;4:1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson C L. Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 41.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 42.Song W, Treich I, Qian N, Kuchin S, Carlson M. Mol Cell Biol. 1996;16:115–120. doi: 10.1128/mcb.16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 44.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]