Abstract
The biochemical characterization of leading and lagging strand DNA synthesis by bacteriophage T4 replication proteins has been addressed utilizing a small, defined primer/template. The ATP hydrolysis activity of 44/62, the clamp loading complex responsible for holoenzyme assembly, was monitored during assembly of both the leading and lagging strand holoenzyme complex. The ATPase activity of 44/62 diminishes once a functional holoenzyme is assembled on both the leading and lagging strand. The assembly of the lagging strand holoenzyme is facilitated by several factors including biotinylated streptavidin blocks at the end of the fork strands, preassembly of the leading strand holoenzyme, and by the presence of the DNA primase with ribonucleoside triphosphates. The resultant minimal replicative complex consists of two holoenzymes and a primase nested on a model replication fork derived from a 62-mer template/34-mer primer/36-mer lagging strand in an apparent 2:2:1:1 ratio of 45 protein:polymerase:primase:forked DNA. The 44/62 protein complex does not remain associated with the complex. The primase alone slowly synthesizes pentaribonucleotides on the forked DNA when the lagging strand contains a nonannealed TTG initiation site with the rate of synthesis greatly stimulated by the addition of the 41 helicase. The addition of deoxy-NTPs to this complex results in leading strand synthesis, but extension of the synthesized RNA primer does not occur. DNA synthesis in both the leading and lagging strand directions is achieved, however, when a 6-mer DNA primer is annealed to the primase recognition site of the forked DNA substrate. A model is presented that describes how leading and lagging strand DNA synthesis might be coordinated as well as the associated molecular interactions of the replicative proteins.
Keywords: DNA replication/DNA polymerase/accessory proteins/DNA primase
Replication of genomic DNA is a complex endeavor involving a confederation of distinct enzyme activities. Bacteriophage T4, one of the more simple DNA replication systems, utilizes eight proteins in the formation and propagation of the DNA replication fork. At the core of the replication process is the T4 DNA polymerase which is responsible for catalyzing nucleotide incorporation while its 3′ → 5′ exonuclease activity acts to maintain fidelity during DNA replication (1). As is the case for most DNA polymerases, the T4 enzyme alone will incorporate nucleotides in a distributive manner as opposed to the highly processive manner required for genomic replication in vivo (2). Through its interaction with replicative accessory proteins that prevent the polymerase from rapidly dissociating from the DNA, the processivity of the T4 polymerase is increased (3). These accessory proteins include the 45 protein, generically referred to as the “sliding clamp” and the 44/62 protein complex that places the sliding clamp onto DNA in an ATP-dependent manner (4). DNA helicase [gene product (gp) 41 (gp41)] is responsible for ATP- or GTP-dependent unwinding of the duplex DNA that is to be replicated (5) while the single-stranded DNA-binding protein (gp32) functions to prevent reannealing of the replication fork (6). The activity of the helicase is enhanced substantially by its own accessory protein, gp59, that appears to act as an assembly factor for the DNA helicase (7). Associated with the helicase is the DNA primase (gp61) which provides the pentaribonucleotide primer needed for the initiation of lagging strand DNA synthesis (8).
The mechanism and dynamics of leading strand DNA synthesis have been addressed kinetically and structurally (reviewed in ref. 9). The holoenzyme assembly process proceeds by at least two ordered events in which the 45 protein is loaded onto DNA in an ATP-dependent manner by the 44/62 protein complex followed by the rapid association of the polymerase with the loaded 45 protein to form the holoenzyme. The 45 protein possesses a ring-shaped structure with an interior diameter large enough to circumscribe duplex DNA (J. Kuriyan, personal communication). Although never directly demonstrated, the 45 protein probably confers enhanced stability upon the holoenzyme through its concatenation with DNA. As such, the 44/62 protein acts as the “clamp loader” to presumably open the ring-shaped 45 protein as well as chaperone the 45 protein to the primer/template. This clamp-loading process is ATP-dependent and the ATP hydrolysis event associated with clamp loading is the rate-limiting step for holoenzyme formation (≈1 sec−1) (10). Once the 45 protein is placed onto DNA, the T4 polymerase rapidly associates under diffusion control with them to form the holoenzyme in which the 45 protein interacts with the carboxyl terminus of the polymerase (11). The 44/62 protein performs holoenzyme formation in a catalytic manner and is not stably associated with the leading strand holoenzyme during DNA replication. Once the holoenzyme is formed, it is relatively stable with a dissociation rate constant (koff) of 0.01 sec−1 (12). This rate of dissociation for the leading strand holoenzyme is sufficiently slow that, given a polymerization rate of approximately 500 nt/sec, processive replication of the T4 genome during a single pass of the holoenzyme can be accomplished.
All prokaryotic and eukaryotic DNA polymerases to date, including the bacteriophage T4 DNA polymerase, share the same fundamental type of synthetic activity in which the DNA primer is extended by the sequential addition of nucleotides to the 3′-OH of a suitable DNA or RNA primer. In vivo, this necessitates that lagging strand DNA replication proceeds in a discontinuous fashion. During DNA replication, the leading strand replication complex processively replicates genomic material to yield large duplex DNA products (>50 kb). The lagging strand, however, is replicated in smaller sections (≈1–2 kb) known as Okazaki fragments (13). The discontinuous nature of lagging strand DNA replication thus poses several intriguing challenges for the replication machinery of any organism.
The lagging strand replication complex must be processive enough to replicate 1–2 kb of DNA, yet must rapidly dissociate once the Okazaki fragment is completed and then recycle itself to a newly formed RNA primer. This recycling problem can be alleviated by the proper disassembly of the lagging strand holoenzyme, but this process must occur in a highly regulated fashion so as not to disturb continuous leading strand DNA synthesis. A partial answer lies in the mechanism by which the lagging strand holoenzyme can disassemble in a timely fashion. Kinetic measurements examining the stability of the holoenzyme once it encounters a completed Okazaki fragment indicate that the holoenzyme dissociates rapidly (koff = 0.3 sec−1) upon encountering the next Okazaki fragment, suggesting that the 5′-triphosphate of the RNA segment is a signal that triggers the decomposition of the holoenzyme (14).
As yet another unresolved conundrum with respect to lagging strand DNA synthesis is the nature and timing of lagging strand holoenzyme assembly, its interaction with the primase to initiate lagging strand synthesis and with the leading strand holoenzyme to coordinate DNA replication. In this paper, kinetic evidence is presented for the formation of leading and lagging strand DNA holoenzymes as a 1:1 complex at a synthetic replication fork. Stoichiometric formation of this complex is dependent on the presence of the DNA primase, [ribonucleoside triphosphates (rNTPs)], and a primase recognition site. Although the DNA primase is capable of synthesizing pentameric ribonucleotides at this model replication fork, the amount and rate of RNA synthesis by the primase alone is not sufficient to support lagging strand DNA synthesis by the T4 polymerase. DNA synthesis in both leading and lagging strand directions, however, is observed with a hexamer DNA bound at the primase recognition site, thus demonstrating functionality of the dimeric replication complex. The requirements for several distinct proteins and the kinetic ordering of their interactions at the replication fork reveal a complex pathway for the assembly of the replicative complex.
MATERIALS AND METHODS
Materials.
Reagents. [γ-32P]ATP, [α-32P]ATP, [α-32P]CTP, and [α-32P]dCTP were purchased from New England Nuclear. Unlabeled deoxynucleoside triphosphates (dNTPs) were obtained from Pharmacia (ultrapure). All oligonucleotides, including those containing biotin derivatives, were synthesized using an Expidite DNA synthesizer according to established protocols. All DNA substrates, either single-stranded or duplex, were purified as previously described (15). Preparation and quantitation of the Bio-34/62/36-mer DNA substrates were performed as before (ref. 10 and references therein). ATP, NADH, phosphoenol pyruvate, Mg(OAc)2, and all buffers were from Sigma. T4 polynucleotide kinase was from United States Biochemical. All other materials were obtained from commercial sources and were of the highest available quality.
Enzymes.
The T4 exonuclease-deficient polymerase D129A (Asp-219 to Ala mutation) was purified as before (16). Both the 44/62 protein and 45 protein were purified from overproducing strains obtained from William Konigsberg (Yale University, New Haven, CT) as indicated (17). T4 DNA primase (gp61) and DNA helicase (gp41) were generous gifts from Kevin Raney (University of Arkansas for Medical Sciences, Little Rock, AR).
Assays.
Steady-state ATP hydrolysis measurements of 44/62 activity during holoenzyme assembly were performed as indicated (10). Oligoribonucleotide synthesis by DNA primase was determined by measuring the amount of radioactive-labeled RNA synthesized after electrophoretic separation. A typical reaction used 500 nM DNA primase preincubated with variable concentrations of biotinylated, forked DNA (0.5–5 μM) followed by the addition of 200 μM rNTPs (ATP, CTP, GTP, UTP) and 50 nM [α-32P]rCTP with 10 mM Mg(OAc)2 in the appropriate reaction buffer. At various times, aliquots of the reaction were quenched with 1 N HCl, extracted with phenol/chloroform, and neutralized with 3 M NaOH in 1 M Trizma base. A 10-μl aliquot of the quenched reaction was added to 10 μl of gel loading dye and oligoribonucleotides were separated by 20% denaturing gel electrophoresis followed by analysis by PhosporImaging techniques. Assembly of the leading strand holoenzyme complex was monitored using the strand displacement assay previously described (18). Assembly of the functional leading and lagging strand DNA holoenzymes was performed, in which 250 nM forked DNA substrate (containing the TTG primase recognition site) and 1 μM streptavidin was incubated with 500 nM T4 DNA primase, 10 mM Mg(OAc)2, 1 mM ATP, and 200 μM rNTPs (either labeled or unlabeled) in the appropriate reaction buffer for variable amounts of time to initiate RNA synthesis. To the reaction was then added sequentially 250 nM 44/62, 45, and T4 exo−. After 3 min, an additional 250 nM 45 and T4 exo− was added, followed by addition of 100 μM dNTPs to initiate both leading and lagging DNA synthesis. Aliquots of the reaction were quenched at various times and manipulated as described above to resolve RNA and/or DNA products.
Alternatively, 250 nM forked DNA substrate (containing the TTG primase recognition site) was incubated with 2 μM 6-mer DNA that could anneal to the primase recognition site in the forked strand and 1 μM streptavidin. To the DNA was then added sequentially 250 nM 44/62, 45, and T4 exo− with 10 mM Mg(OAc)2 and 1 mM ATP. An additional 250 nM 45 and T4 exo− was added, followed by addition of [α-32P]dCTP and the remaining dNTPs (with single-stranded DNA trap, 1 mg/ml) to initiate both leading and lagging DNA synthesis. Aliquots of the reaction were quenched at various times and manipulated as described above to resolve DNA products.
RESULTS AND DISCUSSION
Previously, a molecular interaction between the T4 DNA polymerase and its processivity factor, the 45 protein, had been detected by site-directed mutagenesis and subsequent functional studies of the mutant polymerase. Deletion of just six amino acids from the carboxyl terminus of the T4 polymerase abolishes interactions with the processivity factor while leaving the intrinsic biochemical properties of the polymerase unaffected during distributive DNA synthesis (11). The recent crystal structure of the RB69 DNA polymerase, a phylogenic relative of the bacteriophage T4 DNA polymerase, confirms that the six amino acids at the C terminus of the protein form an extruded “tail” that mediates the interaction of the polymerase with the 45 protein (19). Another interesting observation is that the RB69 polymerase possesses a second, previously unrecognized protruding domain encompassing amino acids 499–554 (19), leading to the proposal that the leading strand holoenzyme physically interacts with that on the lagging strand through this region (19).
To address whether a static, dimeric replication complex can be formed on a model replication fork, the ATPase activity of the 44/62 protein during holoenzyme assembly was monitored utilizing defined, forked, DNA substrates (Fig. 1). This ATPase assay has been previously used to monitor and dissect the mechanism of assembly of the leading strand holoenzyme complex (10). In the presence of a 1:1:1 ratio of Bioforked DNA, 44/62, and 45 proteins, the ATPase activity of the 44/62 protein is synergistically stimulated due to loading the 45 protein onto duplex DNA. Upon addition of one equivalent of T4 exo− polymerase, the ATPase activity of the clamp loader protein is substantially diminished due to the formation of a stable holoenzyme complex. The 44/62 protein acts catalytically so that the resultant leading strand holoenzyme complex is composed solely of the polymerase in conjunction with the 45 protein.
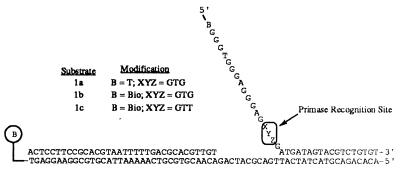
Figure 1.
Substrates used for ATPase and leading and lagging strand DNA synthesis measurements. Modification of DNA with regard to sequence is noted in this figure and described within the text. DNA used to monitor leading and lagging strand DNA synthesis in the absence of DNA primase contained a 6-mer (5′-AACCTC-3′) annealed at the primase recognition site.
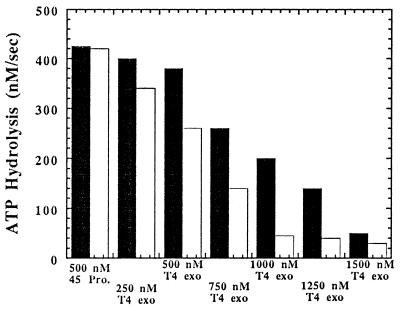
To form a leading and lagging strand holoenzyme complex on a single molecule of DNA simultaneously, the concentration of both the polymerase and the 45 clamp protein must obviously be in twofold excess over DNA and the 44/62 protein. Using 250 nM 44/62, 250 nM Bio-34/62/36-mer (Fig. 1, substrate 1a), and 500 nM 45 protein, a steady-state rate of ATP hydrolysis equal to 420 nM/sec was measured (Fig. 2). Addition of incremental concentrations of T4 exo− decreases the rate of ATP hydrolysis by 44/62 protein, consistent with the formation of holoenzyme complexes. However, at least 1500 nM polymerase is required to diminish ATP hydrolysis by 44/62 protein to its basal rate of 30 nM/sec. This concentration of T4 polymerase corresponds to a high 3:1 molar ratio of T4 exo− to 45 protein and indicates that, under these conditions, formation of a discrete dimeric species is unfavorable.
Figure 2.
Bar graph for the decrease in the ATPase activity of the 44/62 protein associated with the formation of the bacteriophage T4 holoenzyme complex monitoring the decrease in ATPase activity of the 44/62 protein. The concentrations of the 44/62 protein and Bio-34/62/36-mer were maintained at 250 nM while the concentration of 45 protein was fixed at 500 nM. The level of streptavidin was 1 μM while the ATP concentration was fixed at 1 mM. T4 exo− polymerase was incrementally added, and the ATPase activity decreased to eventually reach a limiting rate of 30 nM/sec. The amount of T4 polymerase required to form the leading and lagging strand complexes was reduced in the presence of the biotin-streptavidin barrier (white columns) as opposed to in the absence (black columns), indicating that the formation of the lagging strand holoenzyme is facilitated by the presence of a biotin-streptavidin barrier at the 5′ end of the forked strand.
However, when the identical experiment is performed using a DNA substrate biotinylated at the 5′ end of the forked strand (Fig. 1, substrate 1b), the ratio of T4 to 45 protein required to diminish the ATPase activity of 44/62 protein to its basal rate decreased to 2:1 (Fig. 2). Qualitatively, the lower molar ratio of polymerase to 45 protein implicates the formation of a second holoenzyme that is facilitated by the streptavidin-biotin block at the end of the single-stranded DNA. This result suggests that the 45 protein is capable of diffusing off the end of single-stranded DNA in the absence of streptavidin before the polymerase has the opportunity to bind to the clamp protein and form the holoenzyme complex.
Although these data suggest that leading and lagging strand holoenzymes are being formed on the same DNA molecule, the 2:1 molar ratio of T4 exo− to 45 protein required for this process is not optimal. We anticipate that the physical interaction between the two holoenzymes would provide a tight, stable complex and thus the molar ratio of polymerase to 45 protein in forming both leading and lagging strand holoenzymes should be 1:1. One possibility to account for this discrepancy is that the aformentioned experiments were performed under random assembly conditions in which both leading and lagging strand holoenzymes were assembled simultaneously. However, during DNA replication in vivo, the highly processive leading strand holoenzyme is generally bound in close proximity to the extending primer/template junction. The juxtaposition of the leading strand holoenzyme to that of the lagging strand DNA may provide a “scaffold” that facilitates the formation of the lagging strand holoenzyme.
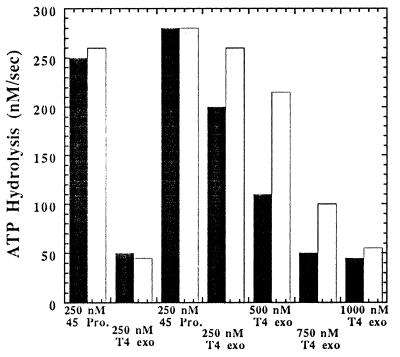
To address this possibility, ATP hydrolysis measurements were performed in which the leading strand holoenzyme was first assembled and then the lagging strand holoenzyme was formed by the sequential addition of 45 protein followed by incrementally increasing concentrations of T4 exo− (Fig. 3). The ATPase activity of the 44/62 protein, using equivalent concentrations of DNA (Fig. 1, substrate 1b), 45 protein, and 44/62 (each 250 nM), was 250 nM/sec. T4 exo− (250 nM) was then added to form the leading strand holoenzyme, after which the ATPase activity of the clamp loader diminished to its basal rate of ≈30 nM/sec. An additional 250 nM 45 protein (that amount required for the lagging strand complex) was then introduced, resulting in a stimulation of ATP hydrolysis by the 44/62 protein to a rate of ≈260 nM/sec. The increase in the rate of ATP hydrolysis by the 44/62 protein again demonstrates the catalytic nature of the clamp loader during holoenzyme assembly, and is also in accord with additional 45 protein being loaded onto DNA already containing a leading strand holoenzyme complex positioned at the primer/template. Following the addition of the clamp protein, the T4 exo− polymerase was then incrementally added until the rate of ATP hydrolysis decreased to the basal rate of ≈30 nM/sec at a 3:1 ratio of T4 exo− to 45 protein. The molar ratio of the polymerase to the processivity factor under ordered assembly conditions suggests that the leading strand holoenzyme forms at a 1:1 ratio whereas the lagging strand holoenzyme assembles at a 3:1 ratio. If, as above, one presumes that the initial holoenzyme is assembled at the leading strand, its formation may influence formation of the lagging strand holoenzyme.
Figure 3.
Bar graph for the formation of the leading and lagging strand holoenzyme complexes under conditions of ordered assembly. The concentrations of Bio-34/62/36-mer, 44/62 protein, and 45 protein were set at 250 nM. Streptavidin was maintained at 1 μM while the ATP concentration was fixed at 1 mM. 250 nM T4 exo− polymerase was added, and the ATPase activity decreased to eventually reach a limiting rate of 30 nM/sec. After formation of the leading strand holoenzyme, 250 nM 45 protein was added followed by incremental additions of the T4 polymerase to form the holoenzyme on the lagging strand. The ratio of T4 polymerase to 45 protein required to form the leading and lagging strand complexes was reduced in the presence of the biotin-streptavidin barrier (black columns) as compared with its absence (white columns).
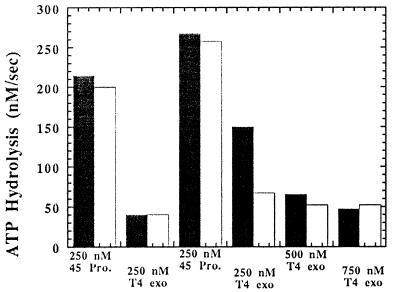
Since DNA polymerases typically do not avidly bind single-stranded DNA or commence polymerization de novo, assembly of either the leading or lagging strand holoenzyme complex followed by synthesis is dependent on the presence of a duplex region. To circumvent this problem at the moving replication fork, pentaribonucleotide primers are synthesized in vivo by the DNA primase at defined sites to which the lagging strand holoenzyme can bind to initiate lagging strand DNA synthesis. According to this in vivo model, the DNA primase may be absolutely required for the in vitro formation of a stoichiometric leading and lagging strand complex at the replication fork. The effect of DNA primase on the formation of the static holoenzyme was addressed in identical fashion to the previously described experiments in which the leading strand holoenzyme was first preassembled followed by the addition of the components for the lagging strand holoenzyme. The DNA substrate in these experiments contained a 3′-TTG-5′ primase recognition site within the forked region of the substrate (Fig. 1, substrate 1c). Addition of 1 μM primase either before or after formation of the leading strand holoenzyme did not facilitate the formation of the lagging strand because identical rates of ATP hydrolysis were measured in the presence or absence of 1 μM primase (Fig. 4). However, when primase and rNTPs are added, the efficiency of forming the lagging strand holoenzyme complex is enhanced as the molar ratio of T4 exo− to 45 protein approaches 1:1 for assembly of both leading and lagging strand holoenzyme complexes (Fig. 4). Furthermore, the presence of a primase recognition site is absolutely required because the molar ratio of T4 exo− to 45 protein remains at 3:1 for assembly of both leading and lagging strand holoenzyme complexes using a forked DNA strand that does not contain a primase recognition site despite the presence of primase and rNTPs (data not shown). Thus, T4 DNA primase, rNTPs, and a usable primase recognition site are the minimal requirements for the formation of a replication complex composed of leading and lagging strand holoenzyme complexes.
Figure 4.
Effect of DNA primase and rNTPs on the formation of the leading and lagging strand holoenzyme complexes. The concentrations of Bio-34/62/36-mer, 44/62 protein, 45 protein, and T4 exo− polymerase were set at 250 nM in the absence or presence of 1 μM DNA primase. The level of streptavidin was set at 1 μM while the ATP and rNTP concentration was fixed at 1 mM and 200 μM, respectively. T4 exo− polymerase was incrementally added, and the ATPase activity decreased to eventually reach a limiting rate of 30 nM/sec. After the addition of 250 nM 45 protein, the ratio of T4 polymerase to 45 protein required to form the leading and lagging strand complexes was reduced in the presence of both DNA primase and rNTPs (white columns) as opposed to in the presence of DNA primase but in the absence of rNTPs (black columns).
The diminution in ATPase activity by the 44/62 protein upon formation of the static replication complex may indicate that the clamp loader remains a stable component of the complex, either associated with the lagging strand holoenzyme and/or mediating the interaction between the leading and lagging strand holoenzymes. The composition of the static replication complex was addressed by determining whether unbound 44/62 or 45 protein remained after assembly of the replication complex. After formation of the static replication complex, ATP hydrolysis was monitored for the case where an aliquot of 250 nM 44/62 protein had been added to the reaction. The lack of enhanced ATP hydrolysis (data not shown) suggests that all 45 protein originally added is sequestered into holoenzyme complexes. However, a large enhancement in ATP hydrolysis was observed when an additional 250 nM 45 protein was added to the solution after formation of the static replication complex (data not shown). The increased ATP hydrolysis indicates that 44/62 is not stably associated with the replication complex similar to its role in the assembly of the leading strand holoenzyme (10).
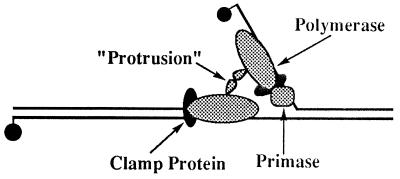
The course of replication complex formation in the presence of primase and rNTPs implicates a physical interaction between the two polymerases and/or 45 proteins that is mediated by the DNA primase (Fig. 5). Several molecular models may account for their tight interaction at the replication fork. In the presence of rNTPs, the primase may synthesize an RNA primer so that the lagging strand holoenzyme can bind to either the RNA primer or to the complex of the primase in association with the RNA. (Note: It is important to recognize that assays monitoring ATP hydrolysis address the formation of stalled leading and lagging strand holoenzymes because dNTPs are omitted.) Alternatively, the addition of rNTPs may stabilize binding of the primase to the recognition site at the forked junction, providing a site for assembly of the lagging strand holoenzyme.
Figure 5.
Proposed model for leading and lagging strand holoenzyme complex formation. Both leading and lagging strand holoenzymes are composed of the T4 polymerase complexed with a 45 “sliding clamp” protein. The formation of a stoichiometric replication complex is dependent on the presence of the T4 DNA primase, rNTPs, and a primase recognition site. The leading and lagging strand holoenzymes are proposed to interact via an internal region protrusion.
To differentiate between these two possible modes as well as demonstrate the functionality of the leading and lagging strand holoenzymes, we monitored nucleotide polymerization complementary to both leading and lagging strand templates. It was first necessary to demonstrate whether the DNA primase would synthesize a pentaribonucleotide primer at this forked DNA junction. If the primase is functional at the replication fork, then the formation of the lagging strand holoenzyme is most likely dependent on RNA synthesis catalyzed by the primase rather than an altered conformation of the primase induced by rNTP binding.
RNA Synthesis Catalyzed by Primase.
DNA primases catalyze the template-directed de novo synthesis of oligoribonucleotides on single-stranded DNA for use as primers to initiate DNA synthesis at origins of replication and at the lagging strand of the replication fork (20). In addition to interacting with the DNA polymerase, the DNA primase also physically and functionally interacts with DNA helicases, in which the primase makes use of the unidirectional translocation activity of the helicase to access the recognition sites for primer synthesis. The bacteriophage T4 DNA primase initiates RNA synthesis from the trinucleotide recognition sequence 3′-T(C/T)G-5′ in which the 3′-nucleotide of the sequence is essential for recognition but is not copied into product oligoribonucleotide (21).
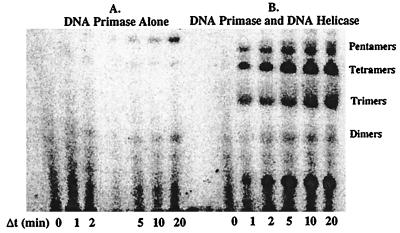
Fig. 6A displays a typical time course for oligoribonucleotide synthesis catalyzed by T4 DNA primase using biotinylated, forked DNA substrate containing a 3′-TTG-5′ recognition site. Appreciable RNA synthesis comprised primarily of pentaribonucleotides (although smaller and larger RNA products are formed) was detected after 20 min, a rate too slow to support lagging strand DNA synthesis in vivo. The rate of RNA synthesis by the T4 DNA primase was stimulated by the addition of an equivalent concentration of DNA helicase (Fig. 6B). A mixture of RNA products from dimers to pentaribonucleotides was observed within 1 min.
Figure 6.
RNA synthesis catalyzed by DNA primase. Pentameric ribonucleotides were produced using forked, biotinylated DNA containing the 3′-TTG-5′ recognition site very slowly by the T4 DNA primase (A). The presence of T4 DNA helicase stimulated the synthesis of RNA primers (B). Assays and quantitation of products are described in Materials and Materials.
Leading and Lagging Strand DNA Synthesis.
Attempts to achieve leading and lagging strand DNA synthesis were performed using stoichiometric levels of DNA and proteins under the conditions of holoenzyme assembly demonstrated by the ATPase assays. The leading strand holoenzyme (250 nM DNA, 44/62, 45, and T4 polymerase) was assembled in the presence of 1 mM ATP and 500 nM primase with 200 μM rNTPs (either labeled or unlabeled). The lagging strand holoenzyme was then assembled by the sequential addition of 250 nM 45 protein and 250 nM T4 polymerase. Following formation of both holoenzymes, dNTPs (either labeled or unlabeled) were added to initiate leading and lagging strand DNA synthesis, and the DNA and/or RNA products were resolved by denaturing gel electrophoresis. We anticipated that once the primase synthesized an RNA primer at the replication fork, the dimeric leading and lagging strand holoenzyme complex would be assembled and poised to synthesize DNA in both directions upon the addition of dNTPs. Only leading strand DNA synthesis, however, was observed under these conditions (data not shown). Most likely, the time required for appreciable pentaribonucleotide synthesis (>10 min) makes extension of the RNA primer by the holoenzyme unfavorable under these conditions. Although pentaribonucleotides are synthesized more rapidly in the presence of DNA helicase, lagging strand synthesis is also not observed when the DNA helicase is present (data not shown). The lack of lagging strand synthesis probably reflects the ability of the helicase, on a small DNA substrate, to rapidly translocate and reload on the forked strand, thus displacing any formed holoenzyme complexes. Utilization of longer DNA substrates should alleviate this problem by preventing the helicase from rapid recycling.
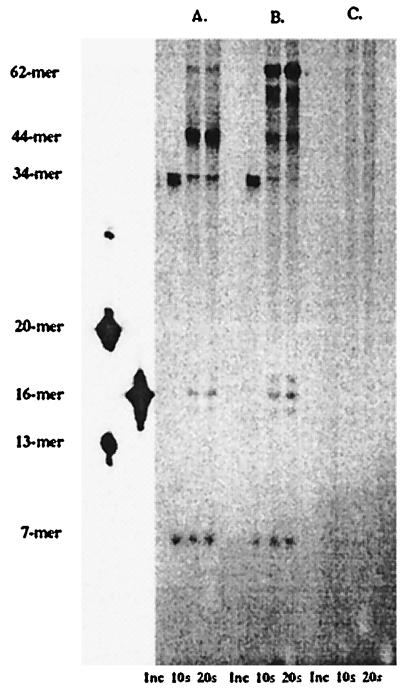
Leading and lagging strand DNA synthesis was monitored in which an artificial lagging strand primer was supplied with the forked, DNA substrate. The primed DNA substrate was formed by incubating 250 nM of the forked DNA substrate (Fig. 1, substrate 1c) with 2 μM of a 6-mer designed to anneal to the primase recognition site. A molar excess of 6-mer DNA was used to force annealing of the small primer to the forked template under these thermodynamically unfavorable conditions. Leading and lagging strand DNA synthesis catalyzed by the T4 DNA polymerase alone was monitored by preincubating the DNA substrate with 750 nM polymerase. Addition of the first nucleotide ([α-32P]dCTP) followed by the remaining dNTPs and a DNA trap allowed for polymerization to occur in both directions. As depicted in Fig. 7A, DNA synthesis occured in both leading and lagging strand directions; leading strand DNA synthesis yielded products greater than 35-mers (using the forked DNA substrate, the T4 polymerase is only able to incorporate nucleotides up to the forked stand, thus yielding products of <44-mers) while smaller DNA products (<20-mers) were also observed, resulting from extension of the lagging strand primer. No extension of the 6-mer primer was observed (Fig. 7C) using 750 nM T4 DNA polymerase incubated with 250 nM of a single-stranded 5′-Bio-36-mer annealed with 2 μM 6-mer. The lack of extension of the short primer/template DNA indicates that the extension products obtained using the forked DNA substrate arise from the lagging strand polymerase binding to the small primer in the confines of the replication fork and extending the small piece of DNA.
Figure 7.
Functionality of leading and lagging strand DNA complexes. DNA synthesis by the T4 polymerase alone occurs in both leading and lagging strand directions with leading strand DNA synthesis yielding products greater than 35-mers while smaller DNA products (16–17-mers) are also observed resulting from extension of the lagging strand (A). The presence of 44/62 protein and 45 protein (B) affects the processivity of the leading strand holoenzyme because strand displacement products are observed (62-mer) and also affects the amount of the lagging strand DNA synthesis. Nucleotide incorporation using 750 nM T4 DNA polymerase with 250 nM of the single-stranded 5′-Bio-36-mer incubated with 2 μM 6-mer (C) was not observed.
The inclusion of accessory proteins (44/62 and 45 protein) enhanced formation of the leading strand holoenzyme complex as strand displacement DNA synthesis was observed (Fig. 7B, DNA products = 62-mer). The accessory proteins also appeared to affect the distribution of lagging strand synthesis products, increasing the amount of the full-length product.
Conclusions.
We have demonstrated the formation of a dimeric leading and lagging strand holoenzyme complex on a defined DNA substrate. Formation of the static complex is facilitated by placing a streptavidin-biotin barrier at the 5′ end of the forked, lagging strand, allowing the polymerase to bind to the 45 protein constrained on the single-stranded DNA. Assembly of the leading strand holoenzyme may facilitate the formation of the lagging strand complex.
A high-affinity replication complex where the two holoenzymes and primase are in a 1:1:1 stoichiometry can be assembled that is dependent on the presence of rNTPs and a primase recognition site. The clamp loader, the 44/62 protein complex, does not appear to remain associated with either the leading or lagging strand holoenzyme and presumably does not participate in DNA elongation or termination of DNA synthesis. Thus, the role of the 44/62 protein is confined to placing the clamp protein onto DNA and to chaperoning the polymerase into a productive interaction with the DNA-bound clamp protein.
At the recognition site on forked DNA, the primase synthesizes pentamers which cannot be efficiently utilized by the DNA polymerase for further elongation of the Okazaki fragment. Hence, although a stoichiometric complex of leading and lagging strand holoenzyme tethered by the DNA primase is formed, it is not functional probably because the primer is sterically inaccessible to the polymerase. The T4 polymerase cannot extend the 6-mer primer annealed to single-stranded DNA although primer extension is observed when the 6-mer is annealed to the same DNA substrate but at the replication fork. This result is consistent with a dimeric replication complex with holoenzymes interacting through a specific domain in the polymerase.
Acknowledgments
This work was supported in part by National Institutes of Health Grant GM13306 (to S.J.B.).
ABBREVIATIONS
- Bio
biotin
- dNTP
deoxynucleoside triphosphate
- rNTP
ribonucleoside triphosphate
- gp
gene product
References
- 1.Alberts B M, Frey L. Nature (London) 1970;227:1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- 2.Nossal N G, Peterlin B M. J Biol Chem. 1979;254:6032–6040. [PubMed] [Google Scholar]
- 3.Huang C-C, Hearst J E, Alberts B M. J Biol Chem. 1981;256:4087–4094. [PubMed] [Google Scholar]
- 4.Mace D C, Alberts B M. J Mol Biol. 1984a;177:279–293. doi: 10.1016/0022-2836(84)90457-1. [DOI] [PubMed] [Google Scholar]
- 5.Nossal N G. J Biol Chem. 1979;254:6026–2031. [PubMed] [Google Scholar]
- 6.Cha T A, Alberts B M. J Biol Chem. 1989;264:12220–12225. [PubMed] [Google Scholar]
- 7.Morrical S W, Hempstead K, Morrical M D. J Biol Chem. 1994;256:4087–4094. [PubMed] [Google Scholar]
- 8.Liu C-C, Alberts B M. Proc Natl Acad Sci USA. 1980;77:5698–5703. doi: 10.1073/pnas.77.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sexton D J, Berdis A J, Benkovic S J. Curr Opin Chem Biol. 1997;1:316–322. doi: 10.1016/s1367-5931(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 10.Berdis A J, Benkovic S J. Biochemistry. 1996;35:9253–9265. doi: 10.1021/bi952569w. [DOI] [PubMed] [Google Scholar]
- 11.Berdis A J, Soumillion P J, Benkovic S J. Proc Natl Acad Sci USA. 1996;93:12822–12827. doi: 10.1073/pnas.93.23.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaboord B F, Benkovic S J. Curr Biol. 1995;5:149–157. doi: 10.1016/s0960-9822(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa T, Okazaki T. Annu Rev Biochem. 1980;49:421–457. doi: 10.1146/annurev.bi.49.070180.002225. [DOI] [PubMed] [Google Scholar]
- 14.Carver T E, Sexton D J, Benkovic S J. Biochemistry. 1997;36:14409–14417. doi: 10.1021/bi971423p. [DOI] [PubMed] [Google Scholar]
- 15.Capson T L, Peliska J A, Kaboord B F, Frey M W, Lively C, Dahlberg M, Benkovic S J. Biochemistry. 1992;31:10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 16.Frey M W, Nossal N G, Capson T L, Benkovic S J. Proc Natl Acad Sci USA. 1993;90:2579–2583. doi: 10.1073/pnas.90.7.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nossal N G. J Biol Chem. 1979;254:6026–6031. [PubMed] [Google Scholar]
- 18.Kaboord B F, Benkovic S J. Biochemistry. 1996;35:1084–1092. doi: 10.1021/bi9520747. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Sattar A K M A, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 20.Kornberg A, Baker T. DNA Replication. 2nd Ed. New York: Freeman; 1992. pp. 113–217. [Google Scholar]
- 21.Hinton D M, Nossal N G. J Biol Chem. 1987;262:10879–10885. [PubMed] [Google Scholar]