Abstract
Tyrosylprotein sulfotransferase (TPST) is a 54- to 50-kDa integral membrane glycoprotein of the trans-Golgi network found in essentially all tissues investigated, catalyzing the tyrosine O-sulfation of soluble and membrane proteins passing through this compartment. Here we describe (i) an approach to identify the TPST protein, referred to as MSC (modification after substrate crosslinking) labeling, which is based on the crosslinking of a substrate peptide to TPST followed by intramolecular [35S]sulfate transfer from the cosubstrate 3′-phosphoadenosine 5′-phosphosulfate (PAPS); and (ii) the molecular characterization of a human TPST, referred to as TPST-2, whose sequence is distinct from that reported [TPST-1; Ouyang, Y.-B., Lane, W. S. & Moore, K. L. (1998) Proc. Natl. Acad. Sci. USA 95, 2896–2901] while this study was in progress. Human TPST-2 is a type II transmembrane protein of 377 aa residues that is encoded by a ubiquitously expressed 1.9-kb mRNA originating from seven exons of a gene located on chromosome 22 (22q12.1). A 304-residue segment in the luminal domain of TPST-2 shows 75% amino acid identity to the corresponding segment of TPST-1, including conservation of the residues implicated in the binding of PAPS. Expression of the TPST-2 cDNA in CHO cells resulted in an ≈13-fold increase in both TPST protein, as determined by MSC labeling, and TPST activity. A predicted 359-residue type II transmembrane protein in Caenorhabditis elegans with 45% amino acid identity to TPST-2 in a 257-residue segment of the luminal domain points to the evolutionary conservation of the TPST protein family.
Protein tyrosine sulfation is a widespread posttranslational modification found in all metazoan species and tissues examined (1, 2). It is catalyzed by tyrosylprotein sulfotransferase (TPST) (3), an integral membrane glycoprotein residing in the trans-Golgi network (TGN) whose catalytic site is oriented toward the TGN lumen (2). Accordingly, proteins trafficking through the TGN have been found to become tyrosine-sulfated, including several identified plasma membrane and secretory proteins (2). As for its physiological role, tyrosine sulfation has been shown to promote protein–protein interaction (2, 4), be it between (i) two secretory proteins (4, 5), (ii) a secretory protein and its cell surface receptor (6, 7), or (iii) two plasma membrane proteins (8–10).
Recognition by TPST requires the occurrence of certain structural features in the substrate protein; a hallmark of these is the presence of acidic amino acid residues in the vicinity of the tyrosine (11–15). The comparison of the sequence motifs of various tyrosine sulfation sites (12), the characterization of the enzymatic properties of TPST from various tissues (16), and the observation that upon SDS/PAGE purified TPST appears as a 54- to 50-kDa doublet of two, albeit highly related, polypeptides (17), have led to the suggestion (2) that for a given organism, isoenzymes of TPST may exist. Here we report the cDNA cloning and molecular characterization of a human TPST, referred to as TPST-2, which is distinct from the human TPST (referred to as TPST-1), whose molecular cloning was reported (18) while this study was in progress.
MATERIALS AND METHODS
Synthesis of [35S]PAPS.
“Carrier-free” [35S]PAPS was synthesized as described (19), purified by thin-layer electrophoresis on cellulose sheets at pH 3.5, eluted, and stored at −20°C. The [35S]PAPS was adjusted to the indicated concentration by addition of unlabeled PAPS.
Synthesis of α,ɛ-Bis-(biotinyl-ɛAhx)-KE(EPEYGE)3-OH (Biotinyl-SgI3).
The synthetic peptide KEEPEYGEEPEYGEEPEYGE [H-KE(EPEYGE)3-OH, referred to as SgI3, identical to SgI-3 (19) except for the presence of one rather than two N-terminal lysine residues] was dissolved in 50% acetonitrile/50 mM sodium borate, pH 8.5, to a final concentration of 3 mM. NHS-LC-biotin (Fluka) in acetonitrile was added from a 30-mM stock solution to a final concentration of 12 mM, and the mixture was incubated for 90 min at room temperature. The pH was maintained at a value of 8.5 by repeated addition of appropriate amounts of 1 M NaOH. After incubation, the mixture was acidified to pH < 3 by addition of trifluoroacetic acid, and α,ɛ-bis-(biotinyl-ɛAhx)-KE(EPEYGE)3-OH (referred to as biotinyl-SgI3) was isolated by using reverse-phase chromatography on a PepRPC 10/10 column (Pharmacia) eluted with a standard acetonitrile gradient, followed by lyophilization. The concentration of biotinyl-SgI3 was calculated from its absorption at 276 nm (ɛ = 4,020 M−1⋅cm−1).
Synthesis of α,ɛ-Bis-benzoylbenzoyl-KE(EPEYGE)3-OH (bzbz-SgI3).
H-KE(EPEYGE)3-OH was dissolved in 50% acetonitrile/50 mM sodium phosphate, pH 7.5, to a final concentration of 3 mM. Benzoylbenzoic acid N-hydroxysuccinimide ester in acetonitrile was added from a 30-mM stock solution to a final concentration of 12 mM, and the mixture was incubated for 90 min at room temperature, with maintenance of pH 7.5 as above. After incubation, α,ɛ-bis-benzoylbenzoyl-KE(EPEYGE)3-OH (referred to as bzbz-SgI3) was isolated by reverse-phase chromatography as described above for biotinyl-SgI3. The presence of the benzoylbenzoyl moiety in bzbz-SgI3 was confirmed and the concentration of bzbz-SgI3 was determined by UV spectroscopy (λmax = 264 nm; ɛ = 28,000 M−1 ⋅cm−1).
Synthesis of α,ɛ-Bis-benzoylbenzoyl-KE(EPEYGE)3YPYDVPYAGK(ɛ-biotinyl-ɛAhx)-amide (bzbz-SgI3-HA-biotin).
α,ɛ-Bis-Fmoc-KE(EPEYGE)3YPYDVPYAGK-amide first was reacted with NHS-LC-biotin as described above for biotinyl-SgI3, and the peptide biotinylated at the ɛ-position of the C-terminal lysine was isolated by reverse-phase chromatography. The Fmoc protecting groups were removed by using 20% piperidine in dimethylformamide. The deprotected peptide was then reacted with benzoylbenzoic acid N-hydroxysuccinimidester as described above for bzbz-SgI3. The α,ɛ-bis-benzoylbenzoyl-KE(EPEYGE)3YPYDVPYAGK(ɛ-biotinyl-ɛAhx)-amide (referred to as bzbz-SgI3-HA-biotin) was isolated by reverse-phase chromatography as described above for biotinyl-SgI3, and its concentration was determined as described above for bzbz-SgI3. The identity of bzbz-SgI3-HA-biotin was confirmed by electrospray mass spectrometry (expected m/z = 4,414, observed m/z = 4,415).
Purification of TPST.
TPST was purified from bovine adrenal medulla as described (17), except that the peptide affinity chromatography was performed by using recombinant sulfophilin (20) instead of the synthetic peptide PKVY+ (17). For this purpose, using standard methods of molecular biology (21), a glutathione S-transferase–sulfophilin fusion protein was expressed in bacteria, purified on glutathione-Sepharose 4B, dialyzed to remove the glutathione, and coupled to Affi-Gel 15 (Bio-Rad) according to the manufacturer’s instructions (82 mg of fusion protein to 20 ml of packed gel, coupling efficiency, 80%).
Three rounds of affinity chromatography were carried out, using a total of 800 ml (200–300 ml per single round) of carbonate-treated membranes, prepared as described (17) from a total of ≈2,000 bovine adrenal medullae. The membranes were solubilized in 70–90 ml of buffer A (ref. 17, except that Mes-NaOH, pH 6.5, was used instead of Tris⋅HCl, pH 7.4) containing 5% (wt/vol) Triton X-100. The supernatant obtained after centrifugation (Triton extract) was supplemented with MnCl2 and nucleotides as described (17), run through a 130-ml PKVY−–Affi-Gel 15 column (17), and the flow-through was loaded onto a 15-ml glutathione S-transferase–sulfophilin–Affi-Gel 15 column. The latter column was washed and TPST was eluted by PAP removal as described previously for the PKVY+–Affi-Gel 15 column (17). Affinity chromatography was monitored by determining TPST activity (ref. 19, using CCK-1 as substrate) and by analyzing aliquots of the eluate by SDS/PAGE and silver staining. Relative to the TPST activity in the Triton extract, ≈11% of the enzyme activity was recovered in the eluate. Analysis of the various fractions of the eluate indicated that the presence of the 54- to 50-kDa doublet thought to represent the TPST protein correlated with TPST activity (data not shown). However, in contrast to the previously reported purification of TPST at the analytical scale (17), the present preparative isolation yielded a few other bands in addition to the 54- to 50-kDa TPST doublet.
The eluates from the three rounds of purification were pooled (80 ml) and proteins were precipitated by addition of polyethyleneglycol 4000 to a final concentration of 20% (wt/vol). To be able to monitor TPST from this step onward, an aliquot of TPST purified as described (17) was labeled with 125I by the chloramine T method, dialyzed, and added as a tracer to the pooled eluates before polyethyleneglycol addition. After incubation on ice for 24 h and centrifugation for 24 h at 93,000 × g, the protein pellet was dissolved in a total of 300 μl Laemmli sample buffer and subjected to SDS/PAGE using a 6% polyacrylamide gel and Tris-Bicine as the running buffer. The gel was fixed and autoradiographed in the wet state at 4°C. The region of the gel containing 125I-labeled 54- to 50-kDa TPST was excised and subjected to trypsin digestion.
Sequencing of Tryptic TPST Peptides.
Gel pieces were washed with water, lyophilized, and rehydrated in 100 μl of 100 mM NH4HCO3/0.5 mM CaCl2 containing 1 μg trypsin (sequencing grade, Boehringer Mannheim). After digestion at 37°C for 12 h, peptides were extracted from the gel with 2 × 100 μl of 70% trifluoroacetic acid in water and 2 × 100 μl 50% trifluoroacetic acid in acetonitrile. The combined fractions were concentrated and subjected to reverse-phase HPLC using Vydac 218TP (2.1 × 250 mm). Automated Edman degradation of peptides was performed by using an Applied Biosystems sequencer (Model 477A) connected to an on-line phenylthiohydantoin analyzer (Model 120) (22, 23).
Cloning of the TPST-2 cDNA.
Standard procedures of molecular biology were used (21). Five micrograms of total RNA, isolated from Caco-2 cells by the guanidine method (21), were reverse-transcribed by using Superscript II RNase H− Reverse Transcriptase (GIBCO/BRL) and (dA,dC,dG)dT18-primers, followed by RNase H digestion. The entire ORF of the human TPST-2 cDNA was amplified by PCR using cloned Pfu polymerase (Stratagene). After identification of the 5′ untranslated region/ORF and ORF/3′ untranslated region boundaries by analysis of the BAC 445C9 and expressed sequence tag (EST) sequences (see Fig. 1B), the 5′ and 3′ primers were designed based on the BAC 445C9 sequence. The 5′ primer was 5′-GGGAAAGCTTCCAGCATGCGCCTGTCGGTGCGGAGG-3′, corresponding to the sequence surrounding the translational start codon (underlined) and introducing a HindIII site (italics). The 3′ primer was 5′-GCTCTAGATTAAATGCATTTTCTTCTCTTCTTGG-3′, corresponding to a sequence ≈30–60 nt 3′ to the translational stop codon and introducing an XbaI site (italics). PCR amplification (3 min at 94°C; 40 cycles, 1 min at 94°C, 1 min at 55°C, 3 min at 74°C; 10 min at 74°C) generated a 1.2-kb fragment, which was cloned into pCR-2.1 (Invitrogen) according to the manufacturer’s instructions, yielding pCR-hTPST2. The entire insert was sequenced. This revealed sequence identity between the PCR-amplified human TPST-2 cDNA and the corresponding parts of the BAC 445C9, except for a C and T at nucleotide position 467 and 898 of the cDNA instead of a G and C in the corresponding position of the BAC 445C9, respectively; these nucleotide substitutions did not alter the protein sequence (A90, L234).
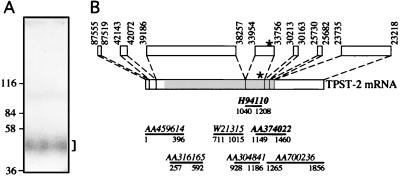
Figure 1.
Purification of bovine TPST (A) and structure of the human TPST-2 gene and mRNA (B). (A) A mixture of TPST isolated on a preparative scale from bovine adrenal medulla by affinity chromatography on glutathione S-transferase-sulfophilin and 125I-labeled TPST purified on an analytical scale as described (17) was subjected to SDS/PAGE followed by autoradiography of the wet gel. The bracket indicates 125I-labeled TPST. The corresponding region of the gel was excised and subjected to tryptic digestion followed by HPLC separation of the peptides and microsequencing. (B) Boxes above the TPST-2 mRNA show the putative exons of the TPST-2 gene in the BAC 445C9; numbers refer to the nucleotides of the BAC 445C9 (Z95115) corresponding to the 5′ and 3′ ends of the exons. The shaded area of the TPST-2 mRNA indicates the ORF. The lines below the TPST-2 mRNA indicate some of the TPST-2 EST clones in the database. Their accession numbers are given above the lines; numbers below the lines give the 5′ and 3′ ends of the EST sequences and refer to the nucleotide sequence of the TPST-2 mRNA. The asterisks and bold accession numbers indicate the exon and EST sequences, respectively, that encode the sequence of the tryptic peptide isolated from purified bovine adrenal medullary TPST.
Northern Blot Analysis.
A multiple tissue Northern blot (CLONTECH) was hybridized at 68°C according to the manufacturer’s instructions, using the 1.2-kb TPST-2 cDNA released from pCR-hTPST2 by EcoRI as a template for generating 32P-labeled probe with the rediprime kit (Amersham). The blot was washed with 2× SSC/0.05% SDS at room temperature and with 0.1× SSC/0.1% SDS at 50°C.
Expression of Recombinant TPST-2 in CHO Cells.
The 1.2-kb HindIII-XbaI fragment from pCR-hTPST2 was cloned into pRC-CMV (Invitrogen), yielding pRC-CMV/hTPST2. CHO cells from one subconfluent 15-cm dish, resuspended in 0.8 ml PBS, were transfected with 50 μg of DNA (pRC-CMV or pRC-CMV/hTPST2) by electroporation (300 V, 960 μF) and replated. After 24 h the medium was changed, sodium butyrate was added to a final concentration of 10 mM, and the cells were incubated for another 17 h and then lysed by scraping in 3 ml per dish of 1% (wt/vol) Triton X-100/100 mM NaCl/1 mM EDTA/10 mM Tris, pH 7.5/10 μg/ml leupeptin/0.25 mM phenylmethylsulfonyl fluoride on ice. The lysate was cleared by centrifugation at 4°C for 10 min at 14,000 × g, and the supernatant was used for determination of TPST activity and peptide crosslinking.
TPST Assay.
For the determination of TPST activity in CHO cell lysates, a novel TPST assay using the biotinyl-SgI3 peptide was employed. The reaction mixture consisted of, in final concentrations, 10 μM biotinyl-SgI3, 2 μM [35S]PAPS (0.4 TBq/mmol), 1% Triton X-100, 5 mM MnCl2, 10 mM MgCl2, 10 mM NaF, and 50 mM Mes-NaOH, pH 6.5. The reaction was started by adding a 20-μl aliquot of CHO cell lysate to a final reaction volume of 50 μl and by placing the sample at 30°C, and was terminated after 1 h of incubation by heating the sample for 1 min to 95°C. Precipitated protein was removed by centrifugation at 14,000 × g, and 40 μl of the supernatant was mixed with 40 μl of a slurry containing 10 μl of streptavidin-agarose (Sigma) in water. The sample was incubated for at least 1 h at room temperature with gentle mixing. The streptavidin-agarose was washed sequentially using 1% Triton X-100/50 mM NaCl/50 mM Tris⋅HCl, pH 7.4; 0.1% Triton X-100/300 mM NaCl/50 mM Tris⋅HCl, pH 7.4; and 0.1% SDS/50 mM NaCl/50 mM Tris⋅HCl, pH 7.4. Radioactive sulfate incorporation into the biotinyl-SgI3 peptide bound to the streptavidin-agarose then was quantitated by liquid scintillation counting. For each CHO cell lysate, TPST activity was determined in triplicate.
Labeling of TPST by Sulfation of a Crosslinked Substrate Peptide.
Unless indicated otherwise, all steps were performed at 4°C. Carbonate-treated membranes, prepared from bovine adrenal medulla as described (17), were processed according to one of the following three protocols. (i) Membranes were solubilized in 2% (wt/vol) Triton X-100/0.1 M NaCl/10 mM MnCl2/10 mM MgCl2/10 mM NaF/50 mM Tris⋅HCl, pH 7.4, to a final protein concentration of ≈5 mg/ml. After incubation for 30 min, insoluble proteins were removed by centrifugation for 10 min at 14,000 × g, and the supernatant was used for peptide crosslinking. (ii) Membranes were solubilized in 0.5% (wt/vol) octylglycoside/0.3 M NaCl/50 mM Tris⋅HCl, pH 7.4, to a final protein concentration of ≈2.5 mg/ml. After incubation and centrifugation as above, the supernatant was supplemented with 5 mM MnCl2/5 mM MgCl2/5 mM NaF and used for peptide crosslinking. (iii) Membranes were solubilized in 1% (wt/vol) Triton X-100/0.3 M NaCl/50 mM Tris⋅HCl, pH 7.4, to a final protein concentration of ≈0.5 mg/ml. After incubation for 1 h, insoluble proteins were removed by centrifugation for 1 h at 140,000 × g, and the supernatant was mixed 1:1 with 40% (wt/vol) polyethylene glycol (average molecular weight, 8,000) and kept overnight. Precipitated proteins were collected by centrifugation for 1 h at 140,000 × g, solubilized in 0.1% (wt/vol) octylglycoside/0.2 M NaCl/5 mM MnCl2/5 mM MgCl2/5 mM NaF/50 mM Tris⋅HCl, pH 7.4, cleared of insoluble material by centrifugation for 30 min at 14,000 × g, and then used for peptide crosslinking.
In the case of CHO cells, the cleared lysate was mixed 1:1 with 40% (wt/vol) polyethylene glycol and kept overnight. Precipitated proteins were collected by centrifugation for 20 min at 14,000 × g, solubilized in 0.1% (wt/vol) octylglycoside/5 mM MnCl2/10 mM MgCl2/10 mM NaF/50 mM Mes-NaOH, pH 6.5, cleared of insoluble material by centrifugation for 10 min at 14,000 × g, and then used for peptide crosslinking.
The extracts received, in a final reaction volume of 100 μl, the bzbz-SgI3 peptide (100 μM) or bzbz-SgI3-HA-biotin peptide (5–10 μM) and carrier-free [35S]PAPS (1–5 × 107 cpm/ml) with or without PAP as indicated in the figure legends, and then were subjected to UV-induced crosslinking for 30–60 min at 4°C by using an Osram HBO 50 HPLC mercury lamp. The light was filtered to remove wavelengths shorter than 310 nm and was focused on the sample by using a Zeiss microscope lamp housing equipped with an adjustable reflector/condenser system. Non-UV-irradiated controls were kept on ice in the dark. Samples then were incubated at 0°C, 30°C, or 37°C as indicated in the figure legends, followed by precipitation of proteins using TCA or acetone. 35S-labeled proteins were analyzed by SDS/PAGE under reducing or nonreducing conditions (±50 mM DTT) followed by transfer to poly(vinylidene difluoride) (PVDF) membranes and autoradiography using a PhosphorImager. In some cases when the bzbz-SgI3-HA-biotin peptide had been used, proteins on the PVDF membrane after autoradiography were immunolabeled according to standard procedures, using the mouse mAb 12CA5 (1:500 dilution, Boehringer) followed by horseradish peroxidase-conjugated secondary antibody (Dianova, Hamburg, Germany) and the enhanced chemiluminescence system (ECL; Amersham).
Database Searches and Computer Analyses.
Nucleotide and peptide sequence databases were searched at the National Center for Biotechnology Information by using the Advanced blast network service (24). The putative exons in the BAC 445C9 encoding the TPST-2 mRNA were identified by comparing the exons predicted by the genscan program (25) with the human EST sequence database and the BAC 445C9 sequence. The predicted protein sequences of human TPST-1 and TPST-2 were aligned by using the multalin 5.3.3 program (26).
RESULTS AND DISCUSSION
Protein Sequence of a Tryptic Fragment Derived from Purified TPST and Identification of the TPST-2 Gene.
To obtain protein sequence information about TPST, the enzyme was purified on a preparative scale from bovine adrenal medulla by affinity chromatography (17) using the substrate sulfophilin, an artificial protein consisting of 12 repeats of the heptapeptide tyrosine sulfation site of chromogranin B (20). After digestion of the TPST band (Fig. 1A) by trypsin and separation of the resulting peptides by reverse-phase HPLC, Edman sequencing of a tryptic peptide yielded the sequence X-X-Y-X-P-Y-A-N/V-P-P-N-Y. Database searches revealed the presence of this sequence (with N in position 8) in two proteins. First, it occurs, preceded by a tryptic cleavage site (see Fig. 2, asterisks), in several human and mouse ESTs shown to encode TPST-1 (18) while this study was in progress. Second, the sequence Y-X-P-Y-A-N-P-P-N-Y occurs in several human and mouse ESTs predicting a protein whose sequence is homologous, but not identical, to human and mouse TPST-1, respectively. This TPST-1-related sequence also occurs, using either the human TPST-1 protein sequence (18) or the sequence Y-X-P-Y-A-N/V-P-P-N-Y in database searches, in the BAC 445C9 (accession number Z95115), which contains a 131-kb human genomic DNA fragment derived from chromosome 22 (22q12.1). Since human TPST-1 maps to chromosome 7 (accession number AF038009), these observations suggested that the BAC 445C9 contains a portion, or all, of a second gene for TPST, referred to as TPST-2 gene.
Figure 2.
Comparison of the amino acid sequence of human TPST-1 (18) and TPST-2. Identical residues are shaded. Boxes indicate residues proposed (18) to be involved in PAPS binding. Bold, solid line, putative transmembrane segment of human TPST-2; triangles, exon boundaries for human TPST-2; asterisks, sequence of the tryptic peptide isolated from purified bovine adrenal medullary TPST.
Organization of the TPST-2 Gene and mRNA.
The BAC 445C9 was analyzed for putative exons, and the information obtained was used to compile the sequence of the putative TPST-2 mRNA from overlapping EST sequences (Fig. 1B). The predicted ORF was corroborated by sequencing a TPST-2 cDNA obtained by reverse transcription–PCR amplification of RNA isolated from Caco-2 cells, a human colon adenocarcinoma cell line.
The human TPST-2 gene consists of at least seven exons, extending over >50 kb on the genome, with the ORF being encoded by exons 3–6 (Fig. 1B). The TPST-2 mRNA has a predicted size of ≈1.9 kb, which is in line with the results of Northern blot analysis (see Fig. 5). The 5′ and 3′ untranslated regions are ≈0.2 kb and ≈0.5 kb long, respectively. The ORF comprises 1,131 nt (start at nucleotide 198–200, stop at nucleotide 1329–1331 of the 1,856-nt mRNA shown in Fig. 1B), encoding a protein of 377 aa residues. The nucleotides surrounding the putative translational start codon (nucleotide 6–8 of the reverse transcription–PCR-amplified TPST-2 cDNA) fit the Kozak rules.
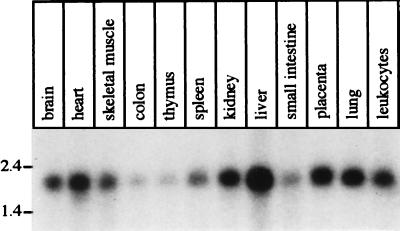
Figure 5.
Northern blot analysis showing the distribution of the TPST-2 mRNA in various human tissues. Each lane contains ≈2 μg of poly(A)+ RNA.
Primary Structure of Human TPST-2.
The primary structure of human TPST-2 predicted from the cDNA is shown in Fig. 2. The TPST-2 polypeptide without posttranslational modifications has a predicted molecular weight of 41,912. Hydropathy analysis revealed a single hydrophobic sequence of ≈18 aa residues sufficient to serve as a membrane anchor, which is located near the N terminus of the protein (Fig. 2, bold, solid line). This finding suggests that TPST-2 is a type II transmembrane protein with a very short N-terminal cytoplasmic tail and the bulk of the polypeptide, which contains two potential N-glycosylation sites (N343 and N368), being located in the Golgi lumen. This would be consistent with the previous observation that TPST is a 54- to 50-kDa integral membrane glycoprotein (17).
The sequence of the peptide obtained from purified bovine TPST is found in both human TPST-1 and TPST-2, but preceded by a tryptic cleavage site only in human TPST-1 (Fig. 2, asterisks). In contrast, both mouse TPST-1 (18) and TPST-2 (mouse TPST-2 EST clones, e.g., AA051791 and AA871253) contain a tryptic cleavage site in the corresponding position. If the same holds true for bovine TPSTs, the TPST peptide presumably would have been derived not only from TPST-1 but also TPST-2 since the TPST purified from bovine adrenal medulla very likely is a mixture of isoenzymes, given the ubiquitous expression of the TPST-1 (18) and TPST-2 (see Fig. 5) mRNAs.
Comparison of human TPST-2 with human TPST-1 revealed an overall 64% amino acid identity between the two proteins (Fig. 2). Most of the variation between the two sequences is found in the N- and C-terminal regions, whereas the segment of TPST-2 from Y64 to Q367 is 75% identical to TPST-1. In contrast to the two potential N-glycosylation sites, all six cysteines in the luminal domain are conserved between TPST-1 and TPST-2. Since the Golgi lumen is thought to contain a thiol-oxidizing milieu and strong thiol-reducing conditions inactivate TPST (data not shown), these cysteines very likely form disulfide bonds, probably within one, rather than between multiple, TPST polypeptide(s) given that recombinant TPST-1 analyzed by SDS/PAGE under nonreducing conditions is largely monomeric (18). The observation that TPST solubilized from membranes and analyzed by SDS/PAGE under nonreducing conditions is at least partially recovered as a disulfide-linked dimer (see Fig. 3) may indicate disulfide bond formation between two TPST polypeptides after membrane solubilization via the single cysteine residue located in the transmembrane segment (Fig. 2). This, in turn, may reflect that native TPST, in line with its sedimentation behavior (17, 27), is a noncovalent homodimer, with dimerization perhaps mediated by the transmembrane segment (28).
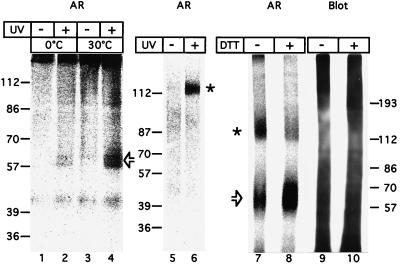
Figure 3.
Identification of TPST from bovine adrenal medulla by MSC labeling. Extracts of carbonate-treated membranes solubilized according to protocol 1 (lanes 1–4), 2 (lanes 5 and 6), or 3 (lanes 7–10) were incubated at 0°C for 30 min (lanes 5 and 6) or 1 h (lanes 1–4, 7–10) with bzbz-SgI3 (lanes 1–4) or bzbz-SgI3-HA-biotin (lanes 5–10) and [35S]PAPS without (lanes 1, 3, and 5) or with (lanes 2, 4, and 6–10) UV-irradiation, followed by further incubation either for 10 min at 0°C (0°C) or 30°C (30°C) (lanes 1–4) or for 1 h at 37°C (lanes 5 and 6) or 30°C (lanes 7–10) in the absence of UV irradiation. Samples were subjected to SDS/PAGE under reducing (lanes 1–4, 8, and 10) or nonreducing (lanes 5–7, and 9) conditions followed by transfer to poly(vinylidene difluoride) membrane and autoradiography (AR, lanes 1–8). The proteins of lanes 7 and 8 then were immunolabeled for crosslinked SgI3-HA-biotin peptide using the mAb 12CA5 against the HA epitope followed by enhanced chemiluminescence (Blot, lanes 9 and 10). Open arrows and asterisks indicate the 61- to 57-kDa TPST monomer and 122- to 114-kDa TPST dimer, respectively, bearing crosslinked [35S]sulfate-labeled SgI3 or SgI3-HA-biotin peptide.
Most of the amino acid residues proposed to be involved in PAPS binding (18) are conserved between TPST-1 and TPST-2 (Fig. 2, boxes). A notable exception is the AKL tripeptide at position 322–324 in TPST-1, which has been suggested (18) to correspond to part of the site in estrogen sulfotransferase (R257-G259) involved in the binding of the 3′ phosphate of PAPS. Whereas in human TPST-1 one of the three residues (K323) is identical to the corresponding residue in estrogen sulfotransferase (K258), the corresponding tripeptide sequence in human TPST-2 is AQL (Fig. 2), which shows no identity to the RKG tripeptide in estrogen sulfotransferase. It thus appears that the binding site for the 3′ phosphate of the cosubstrate in TPSTs differs in amino acid sequence from that of the cytosolic sulfotransferases. It is tempting to speculate that this difference may explain why cytosolic sulfotransferases can be photoaffinity-labeled using [32P]PAPS (29) whereas we have been unable to photoaffinity-label TPST with radioactive cosubstrate (C.N. and W.B.H., unpublished observation).
Identification of TPST by Intramolecular Sulfation of a Photocrosslinked Peptide Substrate (MSC-Labeling).
To identify TPST in crude membrane preparations, we established an approach that is based on the crosslinking of a substrate peptide to TPST followed by intramolecular [35S]sulfate transfer from PAPS. Two synthetic peptides, SgI3 which consists of a triple repeat of the tyrosine sulfation site of chromogranin B (secretogranin I) (19, 30), and SgI3-HA, which contains, in addition, a C-terminal hemagglutinin tag, were modified at the N-terminal lysine by addition of a benzoylbenzoyl (bzbz) group, a photoactivatable crosslinker (31, 32). The bzbz-SgI3-HA peptide was modified further at the C-terminal lysine by the addition of a biotin group. A Triton X-100 extract of carbonate-treated, Golgi-enriched membranes from bovine adrenal medulla was incubated with the bzbz-SgI3 peptide and [35S]PAPS for 1 h on ice, without or with UV irradiation to activate the bzbz crosslinker, followed by further incubation for 10 min at 30°C to enhance [35S]sulfate transfer to the crosslinked SgI3 peptide. SDS/PAGE under reducing conditions followed by autoradiography revealed one major, labeled 61- to 57-kDa doublet that was observed only after UV-induced crosslinking (Fig. 3, lane 4). The apparent molecular mass of this doublet was consistent with the covalent binding of one (or two) ≈4-kDa bzbz-SgI3 peptide(s) to one 54- to 50-kDa TPST polypeptide. (It should be noted that the SgI3 peptide is very acidic, and thus binding of one SgI3 peptide presumably would be sufficient to account for the observed reduction in electrophoretic mobility.) [35S]Sulfate incorporation into the 61- to 57-kDa doublet already was detected when the sample was kept on ice during the crosslinking reaction and thereafter (Fig. 3, lane 2). Since [35S]sulfate incorporation increased 2.5-fold when the 10-min incubation after the crosslinking was carried out at 30°C (Fig. 3, lane 4) rather than 0°C (Fig. 3, lane 2), this increase must have occurred after the covalent binding of the bzbz-SgI3 peptide. This, in turn, suggests that the 61- to 57-kDa protein exhibits TPST activity.
Upon SDS/PAGE under nonreducing conditions, a variable fraction of the molecules bearing crosslinked, [35S]sulfate-labeled SgI3 peptide had an apparent molecular mass of 122–114 kDa (Fig. 3, lanes 6 and 7), consistent with them being covalent homodimers of the 61- to 57-kDa protein. This was corroborated by the observation that treatment with the thiol-reducing agent DTT before SDS/PAGE converted most of the 122- to 114-kDa protein to the 61- to 57-kDa protein (Fig. 3, lane 8).
The specificity of [35S]sulfate-labeling of one major protein after UV-induced crosslinking of the bzbz-SgI3 or bzbz-SgI3-HA-biotin peptide in a relatively crude membrane extract was not a result of selective binding of the SgI3 peptide because immunoblotting with anti-hemagglutinin (HA) antibody revealed proteins bearing crosslinked SgI3-HA-biotin peptide throughout the molecular weight range (Fig. 3, lanes 9 and 10). Rather, the observed specificity of labeling reflected the selective intramolecular [35S]sulfate transfer to SgI3 peptide crosslinked to TPST. If intermolecular [35S]sulfate transfer had occurred, i.e., if TPST had catalyzed the sulfation of SgI3 peptide crosslinked to non-TPST molecules, many labeled bands should have been observed given that the SgI3 peptide was crosslinked to proteins throughout the molecular weight range. It should be noted that the approach of labeling TPST by the transfer reaction from the noncovalently bound labeled cosubstrate to a crosslinked substrate is, in principle, applicable to any transferase. We therefore will refer to this approach with the general term “MSC-labeling” (for Modification after Substrate Crosslinking).
Expression of Recombinant Human TPST-2 in CHO Cells.
To verify that the protein encoded by the human TPST-2 cDNA is indeed a TPST, we expressed it in CHO cells under the control of the cytomegalovirus promoter after butyrate induction, and analyzed the cell lysates by MSC-labeling (Fig. 4A) and by assaying TPST activity (Fig. 4B). For MSC-labeling, the lysate from TPST-2-transfected CHO cells was subjected to photocrosslinking in the presence of the bzbz-SgI3-HA-biotin peptide followed by [35S]sulfate transfer, and analyzed by SDS/PAGE under nonreducing conditions. Only a 125- to 100-kDa protein showed UV-dependent [35S]sulfate-labeling (Fig. 4A Upper), consistent with it being the covalent TPST-2 homodimer. [The greater heterogeneity of the homodimer of human TPST-2 expressed in CHO cells (125–100 kDa, Fig. 4A) compared with that of endogenous TPST from bovine adrenal medulla (122–114 kDa, Fig. 3) is presumably a result of varying glycosylation, a phenomenon frequently observed upon overexpression of proteins in the secretory pathway.] Upon analysis of the same material by SDS/PAGE under reducing conditions, the major [35S]sulfate-labeled band corresponded to the TPST monomer (data not shown). Several other bands were labeled irrespective of UV irradiation (Fig. 4A Upper). These bands reflect cytosolic proteins such as tubulin, which become sulfated by TPST when the membrane boundary between the TGN lumen and the cytoplasm is abolished by detergent (11). The marked increase in the sulfation of these bands in TPST-2-transfected cells as compared with control cells is due to the overexpression of TPST-2. In exposures longer than the one shown in Fig. 4A (Upper), a TPST homodimer bearing crosslinked [35S]sulfate-labeled SgI3-HA-biotin peptide also could be detected in the lysate from control CHO cells transfected with the empty vector, albeit at a 14-fold-lower level compared with TPST-2-transfected cells.
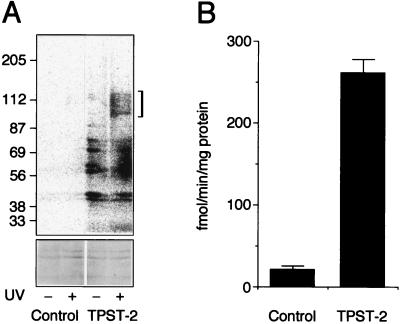
Figure 4.
Expression of human TPST-2 in CHO cells. CHO cells were transfected with pRC-CMV (Control) or pRC-CMV/hTPST2 (TPST-2). (A) MSC labeling. Extracts from the cell lysates (50 μg protein per lane) were incubated for 1 h at 0°C with 10 μM bzbz-SgI3-HA-biotin, 20 μM PAP, and 5 × 107 cpm/ml [35S]PAPS without (−) and with (+) UV irradiation, followed by further incubation for 1 h at 30°C in the absence of UV irradiation. Samples were subjected to SDS/PAGE under nonreducing conditions followed by transfer to poly(vinylidene difluoride) membrane and autoradiography using a PhosphorImager. All four lanes of the top panel are from the same autoradiogram. The bracket indicates the 125- to 100-kDa TPST dimer bearing crosslinked [35S]sulfate-labeled SgI3-HA-biotin peptide. Note the increase in UV-independent [35S]sulfate incorporation into bands of <87 kDa in the TPST-2-transfected cells as compared with control. The two blots in the lower part of the figure show the 87- to 56-kDa region of the Ponceau S-stained poly(vinylidene difluoride) membrane, documenting the presence of equal amounts of protein. (B) Determination of TPST activity in the cell lysate. Data are the mean of two independent transfections; bars indicate the variation of the TPST activity in each cell lysate from the mean.
Determination of TPST activity revealed a 13-fold-greater value in the cell lysate from TPST-2-transfected CHO cells than in that from CHO cells transfected with the empty vector (Fig. 4B). Considering that TPST activity was determined by using the SgI3 peptide as substrate, these results imply that recombinant human TPST-2, like recombinant human TPST-1 (18), recognizes protein substrates bearing the hallmark of tyrosine sulfation sites, the presence of acidic amino acid residues in the vicinity of the tyrosine.
Tissue Distribution of the Human TPST-2 mRNA.
The distribution of the TPST-2 mRNA in various human tissues was investigated by Northern blot analysis of poly(A)+ RNA, using conditions of stringency that should reduce the probability of cross-hybridization of the TPST-2 cDNA probe to the TPST-1 mRNA, given an overall nucleotide identity between the two human ORFs of only 54%. Consistent with the widespread occurrence of tyrosine-sulfated proteins (2, 12), the ≈1.9-kb TPST-2 mRNA was found in all tissues examined (Fig. 5). This is also consistent with the isolation of EST clones for TPST-2 from various human tissues (bone, heart, liver, lung, ovary, placenta, and uterus). Though ubiquitously expressed, TPST-2 mRNA levels in the various tissues were different, being highest in liver and lowest in colon and thymus (Fig. 5). (Reprobing of the Northern blot for actin mRNA confirmed the presence of similar amounts of poly(A)+ RNA for each tissue.) It is worth noting that the expression level of TPST-2 mRNA in the various human tissues is not identical to that reported for TPST-1 (18). For example, TPST-2 mRNA was found to be higher in human liver than in brain (Fig. 5), whereas the converse is the case for TPST-1 mRNA (18). The significance of the different expression levels of TPST-1 and TPST-2 in various tissues remains to be established; perhaps these levels are related to the tissue distribution of the various proteins bearing acidic tyrosine sulfation sites, for which TPST-1 and TPST-2 may exhibit differential specificity.
A TPST Ortholog in Caenorhabditis elegans?
A database search using the human TPST-2 protein sequence revealed the presence of an ORF (F42G9.8, accession number U00051) in the genome of the nematode C. elegans predicting a protein with 45% and 43% amino acid identity to the conserved region of TPST-2 (70–334) and TPST-1 (71–335), respectively. Interestingly, hydropathy analysis of the protein predicted by F42G9.8 would be consistent with it being a type II transmembrane protein. All amino acid residues implicated in PAPS binding and conserved between human TPST-1 and TPST-2 (Fig. 2, boxes, except for the AKL/AQL box) also are conserved in the C. elegans F42G9.8 protein. Furthermore, the F42G9.8 protein exhibits a lower degree of overall amino acid identity to the other Golgi sulfotransferase [19% to chondroitin-6-sulfotransferase (33), 16% to heparan sulfate 2-sulfotransferase (34), 12% to N-heparan sulfate sulfotransferase (35)] than to TPST-1 (33%) and TPST-2 (34%). Given that tyrosine-sulfated proteins have been found in all metazoan species investigated (2, 12) and that TPST hence would be expected to exist in nematodes, we find it likely that the F42G9.8 encodes the C. elegans TPST. If so, this would allow investigation of the general biological significance of protein tyrosine sulfation for an organism by a genetic approach other than inactivating the two TPST genes in mammals identified by Ouyang et al. (18) and in the present study.
Acknowledgments
We thank Dr. Rainer Frank for the synthesis of the unmodified SgI3 peptides and for mass spectrometry of the bzbz-SgI3-HA-biotin peptide, Katja Röper for Caco-2 cell RNA, Ruth Jelinek for help with DNA sequencing, and Yanzhuang Wang for help with the transfection of CHO cells. W.B.H. was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 352) and the European Commission (ERBBIO4CT960058).
ABBREVIATIONS
- TPST
tyrosylprotein sulfotransferase
- MSC
modification after substrate crosslinking
- EST
expressed sequence tag
- HA
hemagglutinin
- PAPS
3′-phosphoadenosine 5′-phosphosulfate
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Huttner W B. Nature (London) 1982;299:273–276. doi: 10.1038/299273a0. [DOI] [PubMed] [Google Scholar]
- 2.Niehrs C, Beisswanger R, Huttner W B. Chem Biol Interact. 1994;92:257–271. doi: 10.1016/0009-2797(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 3.Lee R W, Huttner W B. J Biol Chem. 1983;258:11326–11334. [PubMed] [Google Scholar]
- 4.Huttner W B, Niehrs C, Vannier C. Curr Biol. 1991;1:309–310. doi: 10.1016/0960-9822(91)90094-d. [DOI] [PubMed] [Google Scholar]
- 5.Leyte A, van Schijndel H B, Niehrs C, Huttner W B, Verbeet M P, Mertens K, van Mourik J A. J Biol Chem. 1991;266:740–746. [PubMed] [Google Scholar]
- 6.Mutt V. In: Gastrointestinal Hormones. Glass G B J, editor. New York: Raven; 1980. pp. 169–221. [Google Scholar]
- 7.Rehfeld J F. Int Rev Physiol. 1979;19:291–321. [PubMed] [Google Scholar]
- 8.Wilkins P P, Moore K L, McEver R P, Cummings R D. J Biol Chem. 1995;270:22677–22680. doi: 10.1074/jbc.270.39.22677. [DOI] [PubMed] [Google Scholar]
- 9.Sako D, Comess K M, Barone K M, Camphausen R T, Cumming D A, Shaw G D. Cell. 1995;83:323–331. doi: 10.1016/0092-8674(95)90173-6. [DOI] [PubMed] [Google Scholar]
- 10.Pouyani T, Seed B. Cell. 1995;83:333–343. doi: 10.1016/0092-8674(95)90174-4. [DOI] [PubMed] [Google Scholar]
- 11.Lee R W, Huttner W B. Proc Natl Acad Sci USA. 1985;82:6143–6147. doi: 10.1073/pnas.82.18.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huttner W B, Baeuerle P A. Mod Cell Biol. 1988;6:97–140. [Google Scholar]
- 13.Hortin G, Folz R, Gordon J I, Strauss A W. Biochem Biophys Res Commun. 1986;141:326–333. doi: 10.1016/s0006-291x(86)80372-2. [DOI] [PubMed] [Google Scholar]
- 14.Lin W-h, Larsen K, Hortin G L, Roth J A. J Biol Chem. 1992;267:2876–2879. [PubMed] [Google Scholar]
- 15.Bundgaard J R, Vuust J, Rehfeld J F. J Biol Chem. 1997;272:21700–21705. doi: 10.1074/jbc.272.35.21700. [DOI] [PubMed] [Google Scholar]
- 16.Niehrs C, Huttner W B, Carvallo D, Degryse E. J Biol Chem. 1990;265:9314–9318. [PubMed] [Google Scholar]
- 17.Niehrs C, Huttner W B. EMBO J. 1990;9:35–42. doi: 10.1002/j.1460-2075.1990.tb08077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouyang Y-B, Lane W S, Moore K L. Proc Natl Acad Sci USA. 1998;95:2896–2901. doi: 10.1073/pnas.95.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niehrs C, Kraft M, Lee R W, Huttner W B. J Biol Chem. 1990;265:8525–8532. [PubMed] [Google Scholar]
- 20.Niehrs C, Huttner W B, Rüther U. J Biol Chem. 1992;267:15938–15942. [PubMed] [Google Scholar]
- 21.Ausubel F M, Brent R, Kingston R E, Moore D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1997. [Google Scholar]
- 22.Fiedler K, Kellner R, Simons K. Electrophoresis. 1997;18:2613–2619. doi: 10.1002/elps.1150181417. [DOI] [PubMed] [Google Scholar]
- 23.Rojo M, Pepperkok R, Emery G, Kellner R, Stang E, Parton R G, Gruenberg J. J Cell Biol. 1997;139:1119–1135. doi: 10.1083/jcb.139.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 26.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niehrs C, Stinchcombe J C, Huttner W B. Eur J Cell Biol. 1992;58:35–43. [PubMed] [Google Scholar]
- 28.Laage R, Langosch D. Eur J Biochem. 1997;249:540–546. doi: 10.1111/j.1432-1033.1997.00540.x. [DOI] [PubMed] [Google Scholar]
- 29.Lee R W, Suchanek C, Huttner W B. J Biol Chem. 1984;259:11153–11156. [PubMed] [Google Scholar]
- 30.Benedum U M, Lamouroux A, Konecki D S, Rosa P, Hille A, Baeuerle P A, Frank R, Lottspeich F, Mallet J, Huttner W B. EMBO J. 1987;6:1203–1211. doi: 10.1002/j.1460-2075.1987.tb02355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiele C, Fahrenholz F. Biochemistry. 1993;32:2741–2746. doi: 10.1021/bi00062a002. [DOI] [PubMed] [Google Scholar]
- 32.Dorman G, Prestwich G D. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 33.Williams K J. Curr Opin Lipidol. 1996;7:U202–U208. doi: 10.1097/00041433-199612000-00016. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi M, Habuchi H, Yoneda M, Habuchi O, Kimata K. J Biol Chem. 1997;272:13980–13985. doi: 10.1074/jbc.272.21.13980. [DOI] [PubMed] [Google Scholar]
- 35.Hashimoto Y, Orellana A, Gil G, Hirschberg C B. J Biol Chem. 1992;267:15744–15750. [PubMed] [Google Scholar]