Abstract
A library of long peptides displayed on the pIII protein of filamentous phage was used in biopanning experiments against several protein targets. We find that a large percentage of phage clones that bind specifically to a target contain peptide-encoding genes that do not have an ORF. Instead, the reading frame is either interrupted by one or more nonsuppressed stop codons, or a post-transcriptional frameshift is needed to account for the expression of the minor phage coat protein pIII. The percentage of frameshifted clones varies depending on the target. It can be as high as 90% for clones specific for soluble forms of certain cytokine receptors. Conversely, biopanning against four mAbs did not yield any frameshifted clones. Our studies focused on one clone that binds specifically to rat growth hormone binding protein (GHBP) yet does not have an ORF. A secondary peptide library containing random mutations of this sequence was constructed and panned against GHBP to optimize and correct the reading frame. In the last round (round two) of panning with this library, none of the phage clones that bound to GHBP had an ORF. However, careful analysis of these clones allowed us to design a synthetic peptide capable of binding to GHBP. The results of this study indicate that ORFs are not required to obtain gene expression of the minor coat protein of filamentous phage and suggest that some ORF− clones may have a selective advantage over the clones having ORFs.
Filamentous phage libraries of random peptides have proven to be an excellent source of ligands for many proteins as well as a rich source of information regarding structure-function of proteins and protein–protein interactions (1). In a typical library, the phage carries a recombinant peptide fused to the minor coat protein pIII, and the DNA encoding the peptide is cloned into the gene III of the phage (2–5). In general, a meaningful panning experiment depends on the ability to deduce the peptide sequence from the DNA sequence of the clone that expresses the peptide.
The simplest design of a phage library has the DNA encoding the peptides cloned into the single gene III of the phage DNA. This results, in principle, in every copy of the pIII protein (three to five copies per phage particle) carrying the peptide. An alternative approach is to clone the peptide-encoding DNA into a vector that only contains the gene III and the phage origin of replication (a phagemid). After introduction into an appropriate Escherichia coli host strain, the phagemid and associated peptide are “rescued” by superinfection with a helper phage that contains a weakened origin of replication. In this approach, protein pIII is produced both from the wild-type helper phage gene and from the phagemid pIII-peptide fusion gene. Additionally, because the helper phage carries a weakened origin of DNA replication, the phage produced from this infection preferentially contain the phagemid DNA. As a result of both wild-type and recombinant pIII production, each phage particle contains an average of less than one copy of the peptide–pIII fusion protein.
The peptide library used in biopanning experiments described in this paper is of the latter type. It displays 40-aa peptides encoded by a randomized DNA sequence placed immediately upstream of gene III in a phagemid vector. The library contains 1.5 × 1010 different phage clones and is an excellent source of peptide binders (6, 7).
An unexpected result has been obtained in a series of biopanning experiments using our library. A large fraction of the phage binders obtained did not have an ORF in the peptide-encoding region of the DNA. Instead, the reading frame was interrupted by one or more nonsuppressed stop codons (UGA or UAA in the TG1 E. coli strain used), or proper translation of the downstream gene III would require a shift to another reading frame.
Unusual translational events are well established. Ribosomes are known to misincorporate amino acids at frequencies as high as 10−4 to 10−3 per codon (reviewed in ref. 8). Ribosomes have been documented to be able to follow alternative paths during translation, such as frameshifting to −1, +1, and +6 reading frames, long-distance ribosomal hopping (+60), and reading through stop codons in both eukaryotic and prokaryotic cells (8–11). The HIV gag-pol gene is a well known system in which a highly efficient (12%) −1 frameshift takes place (12, 13). Frameshifting can be responsible for the regulation of gene expression, as shown for the gene encoding the RF2 transcription termination factor of E. coli (14), and has been shown to be associated with a number of diseases, such as some forms of colon cancer (15), Alzheimer’s disease (16), and hemophilia A (17).
The focus of this paper is the analysis of peptides specific to the growth hormone binding protein (GHBP), a soluble form of the growth hormone receptor. Growth hormone (GH; somatotropin) plays an important role in animal growth and development. It regulates a variety of physiological effects, including linear growth of the animal, lactation, differentiation, and electrolyte balance. The molecular mechanism of these biological effects is based on the binding of hormone to GH receptor (reviewed in 18).
GHs from different species share a significant level of sequence homology. The 190-aa human GH has 64% sequence homology to the 189-aa rat GH. GH binds to a GH receptor, which consists of three domains: an extracellular hormone-binding domain (250 aa), a single-pass transmembrane domain, and an intracellular domain. A soluble form of the extracellular domain occurs naturally in blood as GHBP. The initial binding events of hormone to both membrane-bound GH receptor and soluble GHBP are thought to be analogous. Receptor activation requires simultaneous binding of two GH receptor molecules by one GH (i.e., receptor dimerization) to form a complex wherein the two intracellular domains can initiate the process of signal transduction underlying GH activity (19, 20). The model system used in this paper involves the rat GHBP (21).
Our purpose in working with the GH system is to regulate receptor function with surrogate peptide ligands, which is crucial to developing new therapies for diseases such as acromegaly and dwarfism. These peptides are also of potential importance in structure-function studies of the GH receptor. Additionally, a greater understanding of events and mechanisms leading to frameshifting and to expression of genes that do not have an ORF may have practical importance for deriving effective peptide ligands to many proteins of medical importance.
MATERIALS AND METHODS
Peptide Library and Panning.
A degenerate oligonucleotide template was synthesized to encode a 40-aa peptide. The codon design for the gene in the peptide library was NNK (N = G, A, T, or C, and K = T or G), which codes for all 20 amino acids but only one of three possible stop codons (UAG). This codon is suppressed in the E. coli strain used, TG1 [F′ traD36 lacIq lacZM15 proA+B+ /supE Δ(hsdM-mcrB)5(rk−mk− mcrB−) thi Δ(lac-proAB)], leading to the incorporation of glutamine into the polypeptide chain. The peptides were designed to carry the short FLAG epitope (DYKD) (22, 23) at their amino termini and the E-tag epitope (GAPVPYPDPLEPR) (Pharmacia) at their carboxyl termini. The presence of the E tag allows confirmation by ELISA that protein III originating from the phagemid pCANTAB5E (Pharmacia; GenBank accession no. U14321) is incorporated into the phage coat. The diversity of the library (RAPIDLIB) is 1.5 × 1010 members. A more detailed analysis of this library is described elsewhere (6, 7).
A standard method (6) was used to coat and block all microtiter plates with the following exceptions: (i) Coating of the plates with mouse double minute 2 (MDM2) was done in the presence of 2–5 mM DTT, and the phage were panned in the presence of 300 ng/μl glutathione S-transferase; and (ii) in the panning experiment against GHBP using the original (native) library, the microtiter plates first were coated with the capture antibody mAb 4.3 (24) and were blocked with milk before GHBP was added.
The protocol for ELISA analyses of phage has been described (6). After the wells of a microtiter plate were coated and blocked, phage were allowed to bind to the wells, and the wells were washed. Anti-M13 antibody conjugated to horseradish peroxidase was used to detect phage. A clone was considered “positive” if the absorbance readout of the well was ≥2-fold over background.
H10 Secondary Library.
A 145-base oligonucleotide based on the DNA sequence encoding the H10 peptide was synthesized. The sequence of the oligonucleotide is: 5′-CTACAAAGACTTTCCGTGAGTGTGTTGGAGGGCGCATTTTAGGTCATTAGGTTTGGAGTCGTCGTTTGCAGGAGGAG-GTTGATGGTTGGGTTGCTATTTTGTTGCTGGGGT-AGTCGCATGTGTGGCGGCCGCAGTGTGA-3′. Doped nucleosides (underlined) were mixtures containing 95% of the original nucleoside and equimolar concentrations (1.7% each) of the other three nucleosides.
The amplified DNA fragment was cut with the SfiI and NotI restriction enzymes and was ligated into pCANTAB5E previously cut with the same two restriction enzymes. The ligation product was electroporated into the E. coli strain TG1, followed by phage rescue, which provided the secondary phage library based on the H10 clone (1.0 × 1010 members).
Sources of Receptors.
Rat GHBP was prepared as described (21) and was obtained from William Baumbach (American Cyanamid). The MDM2 protein used in biopanning was a fusion of the 1–115 region of MDM2 to glutathione S-transferase. The MDM2 gene fragment was obtained by PCR from the pHDM1A plasmid (25), was cloned into pGEX-5X-1 (Pharmacia), was expressed in a soluble form, and was purified by affinity chromatography on glutathione agarose (Sigma) in the presence of 5 mM DTT. Soluble domains of cytokine receptors other than GHBP were purchased from R & D Systems. To obtain rat GHBP-His6 receptor, the appropriate gene fragment was obtained by PCR by using a cDNA clone of GHBP (21) as a template and then was cloned into pET22 (Novagen). Expression and protein purification were done as described (21).
DNA Sequencing.
The DNA sequence of each clone was determined by automated sequencing on an Applied Biosystems 373 Sequencer by using rhodamine dye or Big Dye chemistries. Sequencing runs were performed in both directions for each clone, and the sequencing data were analyzed with sequencher 3.0 software (Gene Codes, Ann Arbor, MI). In many instances, multiple sequencing runs of the same clone were performed to reduce the possibility of sequencing artifacts.
Competition ELISA.
Microtiter plates were coated with GHBP and were blocked as described above. Before the addition of phage to plates, synthetic H10 peptide (or BSA) was diluted in PBS and was added to duplicate wells (100 μl/well). After incubation for 1 h at room temperature, the prepared phage were added to each well (100 μl/well) without removing the peptide solution. After incubation for 1 h at room temperature, the wells were washed with PBS and were incubated for 1 h with the anti-M13 antibody conjugated to horseradish peroxidase, and the color was developed and analyzed as described above.
Peptide Synthesis.
Synthetic peptide H10 (DYKDLGCYFVAGVVACVKK-biotin) was obtained from a commercial supplier (Anaspec, San Jose, CA). A biotin moiety was coupled to the ɛ-amino group of the C-terminal Lys residue. The peptide was subjected to oxidative conditions after synthesis to allow the formation of a disulfide bond. The peptide was supplied at >80% pure as assessed by HPLC.
BIAcore Analyses.
Recombinant rat GHBP with a His6 tag at the C terminus was immobilized as follows. A solution of 500 nM NiSO4 was injected through two flow cells of an nitrilotriacetic acid sensor chip mounted in a BIAcore X instrument (both from Biacore, Uppsala, Sweden) at a flow rate of 20 μl/min. GHBP at 30 μg/ml in PBSP (20 mM phosphate buffer/150 mM NaCl/0.005% surfactant P-20, pH 7.4) then was injected into one flow cell, and an unrelated His6-tagged single chain antibody was injected into the other flow cell as a control. The injections were completed after 10 min, yielding an increase of 6,000 refractive units.
To measure the binding rate constants of the H10 peptide, nine dilutions of the peptide in PBSP were made (final concentrations of 1–9 μM). Each of these dilutions then was injected across both flow cells for 2 min at a flow rate of 10 μl/min, resulting in refractive unit changes ranging from 50–150 refractive units. The equilibrium dissociation constant (KD) between the H10 peptide and GHBP was calculated by using the steady-state analysis module of the biaevaluation 3.0 software (Biacore). To obtain the dissociation rate constant (Kd), a region of the plot corresponding to the 5- to 10-s post-injection period was analyzed by using the dissociation module of the biaevaluation software.
RESULTS
Phage Library and Panning Results.
The ability to isolate peptide ligands from the phage library was surveyed in a series of biopanning experiments against 12 protein targets, including four mAbs, rat GHBP, six soluble forms (extracellular domains) of cytokine receptors, and MDM2 (a protein ligand of the tumor suppressor p53). Two mAbs, PAb240 and PAb1620, are specific for the tumor suppressor p53. PAb240 recognizes a short linear epitope on p53 (26) whereas the epitope of PAb1620 has been characterized as being conformational (6). The anti-MDM2 mAb 3G5 was recently found to bind to a short linear epitope on MDM2 (27). The epitope of the GH receptor-specific mAb 2C3 (28) was not known.
The panning produced many target-specific phage binders (Table 1). All 18 clones specific for the antibodies had ORFs. However, 74 (56%) of the 133 receptor-specific clones sequenced and tested by ELISA did not have an ORF in the peptide-encoding sequence. Instead, they carried at least one of the nonsuppressed stop codons ochre (UAA) or opal (UGA), or, alternatively, required a frameshift to assure the expression of gene III. Clones without an ORF had a rather balanced distribution with regard to the three different reading frames, with 22, 41, and 38% of the clones in frames 0, +1, and −1, respectively (corresponding to a shift of the reading frame by 0, 1, or 2 nucleotides in the 3′ direction). To further understand the mechanisms involved, we decided to focus on a clone isolated from panning against rat GHBP.
Table 1.
Frequencies of ORF+ and ORF− clones panned from the native library
| Target | ORF+ | ORF− | Total clones | % ORF− | Frame 0 | Frame +1 | Frame −1 | |
|---|---|---|---|---|---|---|---|---|
| PAb 240 | 12 | 0 | 12 | 0.0 | ||||
| PAb 1620 | 2 | 0 | 2 | 0.0 | ||||
| MAb 3G5 | 3 | 0 | 3 | 0.0 | ||||
| MAb 2C3 | 1 | 0 | 1 | 0.0 | ||||
| Total | 18 | 0 | 18 | 0.0 | ||||
| GHBP | 1 | 1 | 2 | 50.0 | 1 | 0 | 0 | |
| IGF-1R | 11 | 4 | 15 | 26.7 | 0 | 4 | 0 | |
| IL-4R | 5 | 14 | 19 | 73.7 | 2 | 8 | 4 | |
| IL-6R | 1 | 7 | 8 | 87.5 | 1 | 3 | 3 | |
| IL-9R | 6 | 5 | 11 | 45.5 | 0 | 3 | 2 | |
| IL-10R | 6 | 20 | 26 | 76.9 | 4 | 7 | 9 | |
| gp130 | 9 | 16 | 25 | 64.0 | 3 | 4 | 9 | |
| MDM2 | 20 | 7 | 27 | 25.9 | 5 | 1 | 1 | |
| Total | 59 | 74 | 133 | 56% | 21.6% | 40.5% | 37.8% |
A set of identical sequences obtained from panning a given target is counted as one clone. All of the clones were shown by ELISA to produce phage that bind to the target with a signal-to-background ratio >2.
The ELISA results suggest that the amount of peptide displayed on the surface of the phage are equivalent between clones with an ORF and clones without an ORF. Many of the clones without an ORF produce ELISA signals equal (or greater) to those with an ORF. Among the clones that are positive for binding to MDM2, the clone that yields the highest signal requires a frameshift to assure expression of gene III. The clone most commonly found among the positives contains an ochre stop codon.
Panning Growth Hormone Binding Protein.
The target protein used in the panning experiments with the native library was the rat GHBP (280 aa), containing the extracellular domain and a hydrophilic tail that results from alternative splicing of the mRNA (18). GHBP was captured with mAb 4.3, which is a non-neutralizing murine IgG specific for the carboxyl terminal tail of GHBP.
A total of 72 clones were picked at random from the second and third rounds of panning and were screened for binding activity. Only 5 round-three clones (5/72) were positive as judged by binding to GHBP, and DNA sequence analyses showed that these were comprised of two distinct clones. The most abundant clone, H10, was present in four of five clones sequenced. The sequence of clone H10 is shown in Fig. 2A. The sequence does not have an ORF as it contains two opal stop codons. Clone H10 binds specifically to GHBP (Fig. 1A), but its binding is not competed by the cognate ligand, that is, bovine GH (Fig. 1B), indicating that clone H10 binds to GHBP at a different site than the GH binding site. In addition, the binding of H10 was not blocked significantly by either the neutralizing mAb 2C3 (28) or the capture mAb 4.3 (data not shown).
Figure 2.
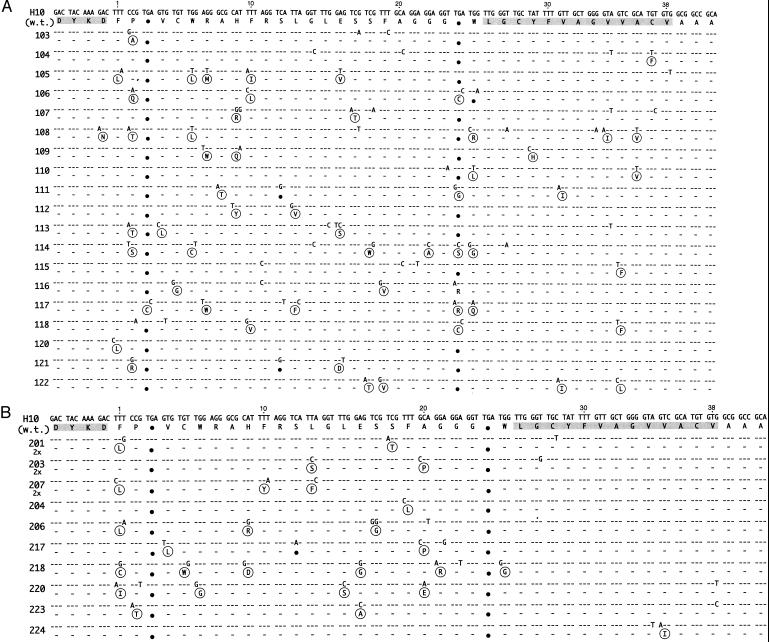
Sequences of H10 mutants. (A) Results of panning round one. (B) Frame 0 sequences from round two. Deduced N-terminal FLAG sequence and the synthetic peptide binder sequence are shaded. Large black dots highlight nonsuppressed stop codons. Sequence differences from the wild-type H10 sequence are shown, and sequence identities are indicated by dashes. Amino acid replacements are shown in circles. The clone number is listed on the left side of the figure. Symbol “2x” under the clone number indicates the number of occurrences of this clone. The UAG codons are translated as Gln in the table because the termination of the translation is suppressed in the E. coli strain used.
Figure 1.
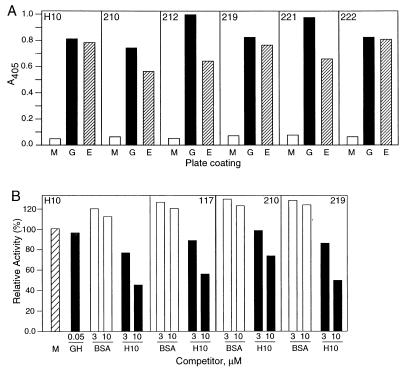
ELISA analysis of phage binders to GHBP. (A) Binding of the H10 phage or its round-two frameshifting mutants to GHBP, anti-E-tag mAb, or nonfat milk-coated plates. Wells were coated with either 2% nonfat milk (M), 100 ng/well of GHBP (G), or 100 ng/well of anti-E-tag mAb (E). Phage were added at 1010/well and were detected by anti-M13 antibody–horseradish peroxidase conjugate. The clone name is indicated in the upper portions of the graphs. (B) Competition of H10 phage binding. Phage were added at 1010/well in the absence or presence of competitor. Competitors were GH, BSA, or the H10 synthetic peptide, added 1 h before phage. Phage were detected by anti-M13 antibody–horseradish peroxidase conjugate. Relative activity is defined as the ratio of ELISA A405 values in the presence and the absence of the competitor. The A405 for the control (no competitor) was 0.4 and is labeled M in the graph.
Secondary Phage Library.
Once the sequence encoding the H10 peptide was established, we constructed a phage library of H10 mutants to search for clones with improved binding properties and to determine whether biopanning would enrich for mutants having an ORF. We reasoned that the best method to generate and to control a high mutation rate per peptide is a gene synthesis-based method in which a synthetic doped oligonucleotide carrying several mutations in any single DNA molecule is used in a PCR to obtain a pool of mutant genes. An average of four mutations were introduced per peptide, which results from a 5% dope rate per nucleoside (calculation not shown). Thus, the number of possible mutant H10 peptide sequences having exactly four mutations (assuming mutations to all other 19 amino acids are possible), 3 × 1011, is close to the anticipated size of the secondary phage library. The mutations were designed to be uniformly distributed along the length of the peptide but exclude the FLAG epitope and the SfiI and NotI restriction enzyme sites. The diversity of the library obtained was 1.0 × 1010 different clones.
Twenty-four clones were randomly selected from the secondary library before panning, and the phage were assayed in an ELISA for binding to the anti-E-tag mAb and to the GHBP (E-tag is used as an indicator of expression of displayed peptides on phage surfaces). The results showed that 46% (11/24) of the clones bind to GHBP, indicating that the maintenance of GHBP binding is a common outcome of this random mutagenesis protocol. Two-thirds of the clones (15/24) were positive in the E-tag ELISA assay. The percentages of GHBP ELISA-positive clones increased, as expected, in rounds one and two to 88% (21/24) and 96% (23/24), respectively. Most clones were E-tag positive by ELISA after rounds one and two, with frequencies of 96% (23/24) and 100% (24/24), respectively. ELISA results for selected clones from round two are shown in Fig. 1A.
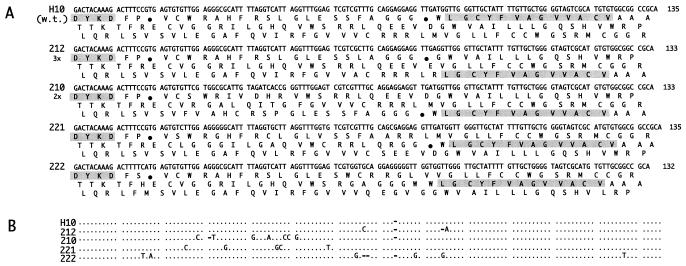
DNA sequences of 34 clones obtained from round one and two are presented in Fig. 2 A and B and Fig. 3 A and B. The clones had an average of 4.2 mutations at the DNA level, resulting in an average of 2.9 mutations at the amino acid level. In round one, despite selection pressure for expression and binding, the nonsuppressed codons were not eliminated in 18 of 19 clones sequenced. Fourteen of 19 clones had at least two nonsuppressed stop codons (clone 121 had three stop codons), two clones had one stop codon, and only one clone, 117, had an ORF. In round two, even this ORF clone was lost. Moreover, in 4 of 14 clones (29%), the reading frame shifted between the FLAG sequence at the 5′ end of the segment and the E-tag sequence at the 3′ end (Fig. 3A). In the rest of the clones, the two original opal stop codons found in the H10 wild-type clone remained (Fig. 3B). One clone, 217, had one ochre and two opal stop codons. A comparison of the results from round one and round two indicates that the ability of the clones to bind was not affected by the positions of stop codons or a shift of the reading frame by either +1 or −1 (compare Fig. 2A with Fig. 3A).
Figure 3.
Sequences of frameshifted H10 mutants (round two). A shows DNA sequences and three reading frames. The FLAG and deduced peptide binder sequences are shaded. Other symbols are as in Fig. 2. B shows a DNA sequence alignment. The alignment compares the H10 sequence with the 221, 222, 212, and 210 sequences. Dots indicate identities, and dashes indicate deletions. The UAG codons are translated as Gln in the figure because the termination of the translation is suppressed in the E. coli strain used.
The distribution of mutations along the polypeptide chain in the round two clones was skewed dramatically toward a high frequency of amino acid substitution in the N-terminal two-thirds of the sequence. In fact, among 26 mutations observed, all except one are located between amino acids 1–25 of the peptides (Fig. 2B). The one exception is a conservative Val to Ile substitution at position 35 in clone 224. The occurrence of nucleotide sequence changes in the frameshifted clones also was biased heavily toward the 5′-terminal two-thirds of the segments (Fig. 3B).
Synthetic H10 Peptide and Its Characterization.
We hypothesized on the basis of mutagenesis results (Fig. 2B) that the sequence conservation between amino acids 26–38 is indicative of the importance of this sequence for target binding, and we designed a peptide encompassing this sequence. The sequence of the synthetic peptide is DYKDLGCYFVAGVVACVKK(biotin).
The binding properties of the synthetic peptide were investigated by ELISA. The synthetic H10 peptide efficiently competed phage-bound peptide clones H10, 210, 117, and 219 in a competitive ELISA assay as shown in Fig. 1B. At a concentration of 10 μM, the peptide reduced the ELISA signal for clones H10, 117, and 219 to 45–55% of the no-peptide control and to 70% of the value for clone 210. GH and BSA had no effect on the binding of the H10 phage to GHBP. These results indicate that the synthetic peptide and the phage-bound peptide bind to the same site on GHBP.
BIAcore analysis of the synthetic H10 peptide–GHBP interaction showed the apparent equilibrium binding constant (KD) to be 2.7 × 10−6 M and the dissociation rate constant (kd) to be 2.6 × 10−2⋅s−1. These values are similar to those reported for other receptor-specific peptides selected from primary phage-displayed libraries (22–25).
DISCUSSION
A typical experiment using a peptide library on filamentous phage relies on biopanning the library against a given target, identifying the phage clones that bind to the target, and sequencing the DNA encoding the peptide. On the basis of the sequence information, a synthetic peptide may be designed, synthesized, and tested for binding to the target. Alternatively, the peptide-encoding gene can be expressed as a protein fusion. Often, it is sufficient to obtain just the sequences of the peptide-encoding genes because the homologies between the peptide sequences (so called “consensus” sequence) may be enough to draw relevant conclusions. It turned out that for a large number of clones from the phage library investigated, none of the three scenarios was possible because of the presence of stop codons, frameshifting, or both in about half of the clones sequenced. (An effort was made to eliminate the possibility of sequencing artifacts, as described in Materials and Methods).
A question arises regarding why the phage library described here, and not any previously published libraries, yielded unusual sequences. Several features distinguish the current phage library. First of all, unlike many other peptide libraries, it is a phagemid-based library; thus, the minor coat protein pIII is present in the cell in two forms: either with a random 40-residue peptide on its N terminus or as the wild-type originating from the helper phage. Phage production and infectivity, therefore, do not depend on the presence of significant amounts of the peptide-pIII fusion; thus, very small amounts of peptide fusion can be incorporated into the phage coat.
Second, the library is mostly composed of long peptides of 40 random amino acids and was constructed from a single 145-nt synthetic oligonucleotide. The consequence of this design is a substantial sequence error rate in the clones from the library, with 46% of the clones in the unselected library not having an ORF because of stop codons or frameshifts (7). Thus, a significant number of unusual sequences were present in the library before panning. The fraction of clones without an ORF was not changed significantly as a result of panning (Table 1).
Third, the nucleotide composition of the clones in the library is skewed toward G and T residues, which comprise 75% of all nucleotide residues (data not shown), rather than the expected 67% based on the NNK codon design. Panning did not affect the nucleotide composition. The high G + T content potentially could lead to a higher propensity of mRNA to form secondary structures caused by G:U base pairing, which, in turn, could affect translation. Despite biases in the nucleotide composition, the amino acid composition is rather balanced and consistent with expectations, with all amino acids found at frequencies varying from 1.3 to 11% (the NNK design gives calculated frequencies of 3 to 10%).
Fourth, the library is one of the largest libraries reported, containing 1.5 × 1010 different clones. Rare clones that would not be seen with smaller libraries can be obtained with a larger library.
It is interesting to note that the frequencies of clones without an ORF strongly depended on the target, with only regular (ORF+) clones being observed for the four mAbs tested whereas the average percentage of ORF− clones for the protein hormone receptors was 56%, the highest percentage being 88% (7 of 8 clones) for the interleukin-6 (IL-6) receptor (Table 1). It may well be that regular clones (with an ORF) are selected preferentially in biopanning experiments, but, if such clones are not available for more difficult targets, biopanning leads to selection of low-abundance clones that do not have an ORF. A possibility also exists that many peptide sequences are toxic to the E. coli host and must be expressed at very low levels. Expression levels from some ORF clones may be too high for cell viability; thus, the selection identifies clones without an ORF. (The lacP promoter driving expression of the gene III fusion in the pCANTAB5E vector and the ribosome binding site native to gene III generally are not used for high level expression in E. coli.)
A study has been undertaken to quantitate the magnitude of the frameshifting effect. We have preliminary data indicating that, for some of the non-ORF and frameshifted segments fused to the E. coli lacZ reporter gene, the expression of β-galactosidase is actually quite high (15–50% of an in-frame control), indicating that successful translation of these clones is not a rare event (data not shown).
A consequence of all three reading frames being able to encode the peptide (Fig. 3A) is that the codon type used can be NNK, as designed, as well as NKN or KNN. In fact, an inspection of the codons encoding the peptide binder (residues 27–38; Fig. 2B) indicates that the codons are of the KNN type. Because of the high G + T content of the sequences, use of KNN codons suggests that Val, Gly, Phe, and Cys residues would be preferred strongly, and, indeed, 9 of 12 residues (shaded region in Figs. 2A and 3A) are residues from this group of four. It is conceivable that the affinity selection retains unusual clones if no regular clones can satisfy the requirement for binding and that, in some cases, an alternative codon design could generate such unusual clones.
A high frequency of frameshifts has been observed in genomic DNA libraries cloned into phage M13. Jacobsson and Frykberg (29, 30) cloned randomly fragmented chromosomal DNA from Staphylococcus aureus into a phagemid vector and subjected the phage to affinity selection for binding to an IgG or to fibronectin. A majority of the resulting clones had the peptide reading frame shifted when compared with either the reading frame for the signal sequence or for protein III. The authors suggest that these findings indicate a translational mechanism involving ribosomal slippage. This same mechanism is a plausible explanation for the frameshifting described in this paper.
A recent paper states that the UGA (opal) stop codon is the least efficient for termination (31). The authors report that a tryptophan is inserted in place of a UGA stop codon in a significant fraction of an overexpressed protein. Substitution of amino acids such as Trp at the UGA and UAA stop codons may be a common event in our peptides. Sequence alignments of MDM2-specific peptides suggest that a Trp is substituted at a UGA codon in at least one clone (data not shown). There also has been one published report of the incorporation of selenocysteine at a UGA codon in a prokaryotic mRNA when the cells are grown under anaerobic conditions (32).
It is possible that a single DNA sequence could be encoding several peptide sequences in the present library because of ribosomal slippage at different sites within the sequence. A single E. coli cell then would produce a heterogeneous population of phage, and the true diversity of the library might be even higher than the nominal diversity of 1.5 × 1010 clones (number of different DNA sequences). The use of such a library would be advantageous when panning against difficult targets, but the benefits of having a phage binder must be balanced against the difficulty of deducing the peptide sequence.
Using one clone specific for GHBP, we have shown that the careful examination of data can lead to the design of a synthetic peptide binder even though no ORF can be identified within the gene. The successful design was based on identifying a “constant” region within the gene: in this case, the latter third (3′ end) of the gene, which is conserved in the clones obtained from a mutational secondary phage library. The specific binding of the peptide to GHBP was demonstrated by ELISA as well as by BIAcore. The same peptide design principle using the sequence near the 3′ end of the gene also has been used successfully for ORF− phage clones specific for two other proteins, IGF1-R and MDM2 (data not shown).
Although the accepted notion is that genes having a frameshift are rare, we find that genes without an ORF are common sources of proteins in clones obtained from the phage library described. This finding illustrates the uncertainty in identifying genes and gene products in complex genomes and in deducing protein sequences from DNA or mRNA sequences. It also emphasizes the level of difficulty in implementing selection schemes in gene libraries of growing complexity.
Acknowledgments
We thank Drs. W. Baumbach and B. S. Wang of American Cyanamid for providing us with rat GHBP and GH and mAbs 2C3 and 4.3. DNA sequencing was done expertly by Darek Galkowski, Denise Steiger, Victor Ferlise, and Ripka Sethi. The MDM2 clone was obtained from Dr. A. J. Levine of Princeton University.
ABBREVIATIONS
- GH
growth hormone
- GHBP
growth hormone binding protein
- MDM2
mouse double minute 2
- IL
interleukin
References
- 1.Scott J K, Smith G P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 2.Grihalde N D, Chen Y C, Golden A, Gubbins E, Mandecki W. Gene. 1995;166:187–195. doi: 10.1016/0378-1119(95)00658-3. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y C J, Delbrook K, Dealwis C, Mimms L, Mushawar I K, Mandecki W. Proc Natl Acad Sci USA. 1996;93:1997–2001. doi: 10.1073/pnas.93.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kay B K, Adey N B, He Y S, Manfredi J P, Mataragnon A H, Fowlkes D M. Gene. 1993;128:59–65. doi: 10.1016/0378-1119(93)90153-t. [DOI] [PubMed] [Google Scholar]
- 5.Lowman H B. Annu Rev Biophys Biomol Struct. 1997;26:401–424. doi: 10.1146/annurev.biophys.26.1.401. [DOI] [PubMed] [Google Scholar]
- 6.Ravera M W, Cárcamo J, Brissette R, Alam-Moghé A, Dedova O, Cheng W, Hsiao K C, Klebanov D, Shen H, Tang P, et al. Oncogene. 1998;16:1993–1999. doi: 10.1038/sj.onc.1201717. [DOI] [PubMed] [Google Scholar]
- 7.Mandecki W, Brissette R, Cárcamo J, Cheng W, Dedova O, Hsiao K C, Moghé A, Ravera M, Shen H, Tang P, Blume A. In: Display Technologies: Novel Targets and Strategies. Guttry P C, editor. Southborough, MA: International Business Communications; 1997. pp. 231–254. [Google Scholar]
- 8.Parker J. Microbiol Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkins J F, Weiss R B, Gesteland R F. Cell. 1990;62:413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atkins J F, Weiss R B, Thompson S, Gesteland R F. Annu Rev Genet. 1991;25:201–228. doi: 10.1146/annurev.ge.25.120191.001221. [DOI] [PubMed] [Google Scholar]
- 11.Weiss R B. Curr Opin Cell Biol. 1991;3:1051–1055. doi: 10.1016/0955-0674(91)90128-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson W, Braddock M, Adams S E, Rathjen P D, Kingsman S M, Kingsman A J. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 13.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Nature (London) 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 14.Craigen W J, Cook R G, Tate W P, Caskey C T. Proc Natl Acad Sci USA. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laken S J, Petersen G M, Gruber S B, Oddoux C, Ostrer H, Giardiello F M, Hamilton S R, Hampel H, Markowitz M, Klimstra D, et al. Nat Genet. 1997;17:79–83. doi: 10.1038/ng0997-79. [DOI] [PubMed] [Google Scholar]
- 16.van Leeuween F W, de Kleijn D P V, van den Hurk H H, Neubauer A, Sonnemans M A F, Sluijs J A, Köycü S, Ramdjielal R D J, Salehi A, Martens G J M, et al. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 17.Young M, Inaba H, Hoyer L W, Higuchi M, Kazazian H H, Jr, Antonarakis S E. Am J Hum Genet. 1997;60:565–573. [PMC free article] [PubMed] [Google Scholar]
- 18.Strobl J S, Thomas M J. Pharmacol Rev. 1994;46:1–34. [PubMed] [Google Scholar]
- 19.de Vos A M, Ultsch M, Kossiakoff A A. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 20.Wells J A. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baumbach W R, Horner D L, Logan J S. Genes Dev. 1989;3:1199–1205. doi: 10.1101/gad.3.8.1199. [DOI] [PubMed] [Google Scholar]
- 22.Knappik A, Plückthun A. BioTechniques. 1994;17:754–761. [PubMed] [Google Scholar]
- 23.Hopp T P, Prickett K S, Price V, Libby R T, March C J, Cerretti P, Urdal D L, Conlon P J. Biotechnology. 1988;6:1205–1210. [Google Scholar]
- 24.Sadeghi H, Wang B S, Lumanglas A L, Logan J S, Baumbach W R. Mol Endicrinol. 1990;4:1799–1805. doi: 10.1210/mend-4-12-1799. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legros Y, Meyer A, Ory K, Soussi T. Oncogene. 1994;9:3689–3694. [PubMed] [Google Scholar]
- 27.Böttger A, Böttger V, Garcia-Echeverria C, Chène P, Hochkeppel H-K, Sampson W, Ang K, Howard S F, Picksley S M, Lane D P. J Mol Biol. 1997;269:744–756. doi: 10.1006/jmbi.1997.1078. [DOI] [PubMed] [Google Scholar]
- 28.Wang B S, Lumanglas A A, Bona C A, Moran T M. Mol Cell Endocrinol. 1996;116:223–226. doi: 10.1016/0303-7207(95)03718-7. [DOI] [PubMed] [Google Scholar]
- 29.Jacobsson K, Frykberg L. BioTechniques. 1995;18:878–885. [PubMed] [Google Scholar]
- 30.Jacobsson K, Frykberg L. BioTechniques. 1996;20:1070–1081. doi: 10.2144/96206rr04. [DOI] [PubMed] [Google Scholar]
- 31.MacBeath G, Kast P. BioTechniques. 1998;24:789–794. doi: 10.2144/98245st02. [DOI] [PubMed] [Google Scholar]
- 32.Chen G T, Axley M J, Hacia J, Inouye M. Mol Microbiol. 1992;6:781–785. doi: 10.1111/j.1365-2958.1992.tb01528.x. [DOI] [PubMed] [Google Scholar]