Abstract
Nuclear hormone receptors comprise a large family of zinc finger transcription factors, some with hydrophobic ligands, such as thyroid hormone, vitamin D, steroids, etc., and others for which no ligand has been found. Thyroid hormone receptors (TRs) generally are considered to be confined to the vertebrata that possess a thyroid gland. Tunicates represent the most primitive of the chordates, and there are data supporting a role for thyroid hormone in their metamorphosis, but no data are available on TRs in this genus; hence, we have studied Ciona intestinalis. Screening of a Ciona library with the DNA binding domain of Xenopus laevis TR (xTR) resulted in the isolation of a nuclear hormone receptor, C. intestinalis nuclear receptor 1 (CiNR1). CiNR1 is similar to TRs of more evolved species with a conserved DNA binding domain whereas the ligand binding domain shows poor homology to vertebrate sequences. The C-terminal part of CiNR1 spans ≈200 amino acids more than other TRs, lacks the AF2 transactivation domain, and is not able to bind triiodothyronine. Phylogenetically, CiNR1 appears to be close to the common ancestral gene of TRs. Expression of CiNR1 was limited to the developing embryo and the larval stage, which suggests a role during development and metamorphosis. In transfection experiments, CiNR1 down-regulated basal transcription of a reporter gene driven by the TR palindrome responsive element. When CiNR1 was cotransfected with chicken TRα, it attenuated the normal thyroid hormone response in a dominant negative fashion. This attenuation required the C-terminal portion of the molecule.
Nuclear receptors are zinc finger transcription factors that regulate gene expression by binding to specific cis-acting sequences in the promoter region (1–3) and include receptors for several hydrophobic ligands, such as steroids, retinoic acid, thyroid hormone, vitamin D, ecdysone, as well as a variety of receptors, called orphans, without a known ligand (4). To date, >150 different members of this superfamily, spanning a large diversity of species from nematode to human, have been described (5).
From the evolutionary point of view, all nuclear receptors can be grouped into six subfamilies: (i) a large one containing thyroid receptors (TRs), retinoic acid receptors (RARs), vitamin D receptors, ecdysone receptors, and numerous orphan receptors; (ii) one clustering retinoid X receptors, COUP, HNF4, and other orphan receptors; (iii) one containing steroid hormone receptors; (iv) one the NGFB group of orphan receptors; (v) one containing FTZ-F1 orphan receptors; and (vi) the last one containing only the GCNF1 orphan receptor (6). Although the TR/RAR and steroid receptor subfamilies are well characterized in vertebrates, little is known about their presence in lower species. Orphan receptors are found in most primitive animals, many of which have homologues or closely related genes in vertebrates (4). The common modular structure of nuclear receptors, their genomic organization, and their conservation in the respective domains suggest that they all evolved from a single ancestor gene (7, 8). The identification of this postulated ancestor could be a tool for clarifying the functional roles of evolved nuclear receptors.
Because there is evidence for thyroid hormone (9, 10) and retinoic acid (11) effects in tunicates, one of the most primitive of the chordate subphylum, we chose the tunicate Ciona intestinalis to investigate the evolution of this subfamily. The position of C. intestinalis in the evolutionary tree is in the point of passage from invertebrate to vertebrate. Its adult body structure is similar to invertebrates, but the tadpole larva has the body plan of vertebrate embryo (12), with a dorsal nervous system, a ventral digestive tract, and a notochord, all of which disappear during metamorphosis. In the present study, we report the isolation in C. intestinalis of CiNR1, a gene that possesses a high degree of homology with other nuclear receptor genes and appears to be close to the ancestor gene of TRs.
MATERIALS AND METHODS
Animals.
Adult C. intestinalis were collected in the Gulf of Naples and were kept in running sea water until they were killed. Ciona embryos and tadpoles were obtained by mixing sperm and eggs from different adult animals. Gametes were obtained by surgical removal from gonoducts. The eggs were put in sea water and then were inseminated with sperm from 2–3 animals to obtain synchronous development. Eggs and sperm were incubated at 18°C with gentle rotation. The incubation was stopped at different times, and the animals at the appropriate stages were collected by low speed centrifugation and were killed for DNA and RNA extraction (13, 14) or were fixed for whole-mount in situ hybridization.
Library Screening.
About 1.5 × 105 recombinant λ phages of a genomic library constructed into EMBL3 SP6/T7 (CLONTECH) with MboI partially digested C. intestinalis muscle DNA were screened with a DNA binding domain (DBD) fragment (839 bp long; PstI digested) of Xenopus laevis TRβA1 obtained through the courtesy of D. D. Brown (Carnegie Institution, Washington, D.C.). Hybridization of phage lifts was carried out at low stringency in hybridization buffer [6× standard saline citrate (SSC)/5× Denhart’s/100 μg/ml salmon sperm DNA/0.1% SDS] containing denatured probe (106 cpm/ml) at 58°C for 14–16 h. Filters were washed once in 2× SSC + 0.1% SDS for 30 min at room temperature and once in 1× SSC + 0.1% SDS for 30 min at 55°C. Recombinant phage DNA was extracted according to Manfioletti and Schneider (16). We obtained two positive clones (λ1 and λ7) with inserts of 12 kilobases, which appeared to be identical, after Southern blot analysis. About 10 μg of phage DNA were digested with EcoRI, and 1 μg of the digested DNA was run in 1% agarose gel and was blotted onto nylon membrane (Hybond-N, Amersham). Filters were hybridized with the same probe used for the library screening. Nine micrograms of EcoRI-digested DNA was run on the agarose gel, and one fragment (3,500 bp) that contained the DBD was purified, was subcloned in pBluescript II SK/KS (+/−) vectors (Stratagene), and then was sequenced. All of the sequences were carried out according to the chain-terminating dideoxynucleotide procedure (17).
A cDNA library (≈5 × 105 phages) from the larval stage constructed in Uni-ZAP XR vector (Stratagene) was screened with the 157-bp-long probe obtained from digestion of λ1 with the restriction enzyme RsaI. The filter was hybridized at high stringency in hybridization buffer with denatured probe (106 cpm/ml) for 14–16 h at 60°C and was washed 4 times in 1× SSC + 0.1% SDS at 60°C for 20 min. Four phages obtained from the last screening were used to infect XL1-Blue bacteria for the excision (Stratagene). The plasmids obtained were tested by digestion with endonuclease enzymes and were sequenced.
Sequence Analysis and Construction of Phylogenetic Tree.
The deduced amino acid sequence was compared with those contained in the GenBank and European Molecular Biology Laboratory database by using the tfasta program from the Genetics Computer Group (GCG) package (18).
DBD amino acid sequences of 18 nuclear receptors from different species were extracted from the GenBank database. Distance matrices were calculated by using the Kimura correction method for multiple substitution in amino acid sequences (19); tree construction was performed by using the growtree phylogram program from Genetics Computer Group package (18).
In Vitro Expression of CiNR1.
A Bluescript plasmid obtained from excision after the cDNA library screening containing the entire ORF of CiNR1 was linearized with XhoI for in vitro transcription with T3 RNA polymerase and subsequently was translated in reticulocyte lysate (Amersham). Labeling of protein was performed with l-[35S]methionine and was analyzed by SDS/PAGE.
Northern Blot.
Total RNA from larval and adult tissues of C. intestinalis was extracted according to Chomczynski and Sacchi (13). The RNA species were separated by electrophoresis on a 1.3% agarose/3% formaldehyde gel and were blotted onto nylon filters (Hybond-N). Blots were hybridized with a 925-bp probe obtained from digestion of CiNR1 with restriction enzymes AvaI and HindIII at 65°C for 14–16 h in hybridization buffer and were washed once with 2× SSC + 0.1% SDS for 20 min at 65°C and once with 0.1× SSC + 0.1% SDS for 20 min at 65°C. Blots were exposed overnight to x-ray film (Kodak). Oligonucleotide sequence against the mouse 28S that cross-reacts with Ciona ribosomal subunit (mouse 28S: 5′-AACGATGAGAGTAGTGGTATTTCACC-3′) was used for comparison in hybridization buffer at 42°C for 14 h and was washed twice with 2× SSC + 0.1% SDS for 20 min at 42°C.
Whole-Mount in Situ Hybridization.
Whole-mount in situ hybridization was carried out according to Caracciolo et al. (20). In brief, cRNA transcribed from either the coding (anti-sense probe) or the noncoding strand (sense probe) of the entire CiNR1 cDNA cloned in Bluescript SK vector, as suggested by the manufacturer (Dig RNA Labeling kit, Boehringer Mannheim), was used. Larvae and embryos were fixed, dehydrated, and stored at −20°C. After rehydration, embryos were dechorionated manually. All of the stages were treated with proteinase K and were postfixed with 4% paraformaldehyde in PBS containing 0.1% Tween 20 for 1 h. Prehybridization was carried out in 50% formamide, 5× SSC, 50 μg/ml heparin, 50 μg/ml tRNA, 5× Denhardt’s solution, and 0.1% Tween for 1 h at 48°C. The prehybridization buffer was replaced with hybridization buffer containing 0.25 μg/ml digoxygenin-labeled antisense or sense transcript.
Hybridization was carried out at 48°C overnight. After the hybridization, larvae and embryos were washed sequentially in 50% formamide, 4× SSC, and 0.1% Tween 20 (2 × 15 min, 48°C); 50% formamide, 2× SSC, and 0.1% Tween 20 (2 × 15 min, 48°C); and sol A (0.5 mol/l NaCl/10 mmol/l Tris, pH 8.0/5 mmol/l EDTA/0.1% Tween 20) (3 × 10 min, 37°C). Digestion with RNaseA (20 μg/ml in sol A) was performed for 30 min at 37°C and was followed by sequential washes with 50% formamide, 1× SSC, and 0.1% Tween 20 (2 × 15 min at 48°C); 1× SSC/PBS containing 0.1% Tween 20 1:1 (15 min); and PBS containing 0.1% Tween 20 (4 × 5 min). After blocking in 5% normal sheep serum in PBS containing 0.1% Tween 20 (30 min), RNA hybrids were detected with 1:2000 alkaline-phosphate-conjugate anti-digoxygenin (Boehringer Mannheim) and were treated for the development of color as indicated in the protocol from Boehringer.
Plasmids and Transfections.
The plasmid 3TRE-pal contains three copies of a palindromic thyroid hormone response element 5′ to a TK-luciferase reporter gene. PSG5cTRα was constructed by cloning the entire coding region of chicken TRα (21) in the pSG5 expression plasmid, downstream of an SV40 promoter. The entire coding region of CiNR1 and the mutated CiNR1 (Mt-CiNR1) gene with a deletion of the last 300 amino acids were cloned in pSG5 expressing vector. COS7 cells were cultured in DMEM supplemented with 10% fetal calf serum. One day before the transfection, cells were plated out in DMEM containing 10% fetal calf serum that was stripped of T3 by ion exchange (22). Cells were transfected by calcium phosphate coprecipitation with 3 μg of reporter plasmid 3TRE-pal, 0.3 μg of vector expressing chloramphenicol acetyl transferase type I (CAT) as an internal control, 1 μg of pGS5cTRα, 6 μg of pSG5CiNR1, and 6 μg of pSG5Mt-CiNR1. For any experiment, the total amount of pGS5 vector, with or without receptor insert, was maintained constant. The precipitated DNA mixture was left on the cells for 16 h. The cells were rinsed in PBS and then were cultured in DMEM with 10% of fetal calf serum that had been stripped of T3. T3 (Sigma) was added at the concentration of 10−7 M. After 2 days, the cells were lysed by adding 100 μl of lysis buffer (10 mM Hepes, pH 7.8./0.1 mM EDTA/5% glycerol/400 nM NaCl/1 mM DTT/1 mM phenylmethylsulfonyl fluoride). Luciferase assay was performed from 3 μl of cell lysates. The luciferase light units were normalized with respect to CAT activity of the cell lysates. The CAT activity was measured by CAT ELISA (Boehringer, Mannheim).
Electrophoresis Mobility Shift Assay.
For binding experiments, 4 μg of cellular extract containing the receptor to be assayed were added on ice in binding buffer (20 mM Hepes, pH 7.9/50 mM KCl/5 mM MgCl2/15% glycerol/0.3 μg/ml poly(dIdC)/5 mM DTT). Then, 32P-labeled PAL oligonucleotide (5′-AATTCTCAGGTCATGACCTGAG-3′) was mixed and incubated for 30 min on ice. Protein–DNA complexes were separated on prerun 5% polyacrylamide/0.25× TBE gel. The gel was dried and autoradiographed.
RESULTS
Cloning of CiNR1 cDNA.
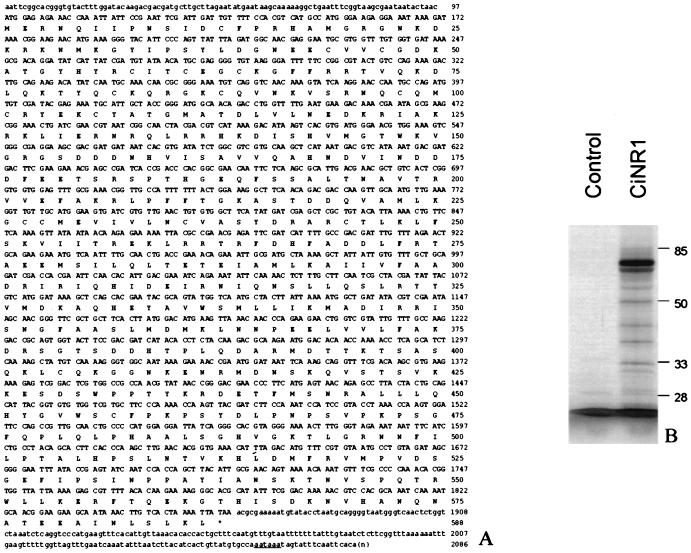
We screened a C. intestinalis cDNA library from the larval stage with a probe that contains the second zinc finger of CiNR1 obtained from the genomic library screening. Four positive clones were identified. Three clones had the same length, and one was shorter, lacking the 5′ portion. The nucleotide and the deduced amino acid sequence of the 2,086-bp cDNA clone named CiNR1 is shown in Fig. 1A. The sequence includes an ORF of 1,761 bp that starts with a putative initiation codon (ATG) at nucleotide 98 and ends with a putative stop codon (TAA) at nucleotide 1,859. The Kozak (23) consensus sequence around the presumed start codon is well conserved. An in frame terminator codon, six codons upstream of the first ATG codon, provides support for the initiation methionine, although the presence of an additional methionine 19 codons downstream makes this assignment tentative. Like many other members of this family (24, 25), the DBD of CiNR1 contains the classical nine cysteine residues, eight of which participate in the formation of two zinc fingers. A typical polyadenylation signal is present 23 nucleotides upstream of the poly-A tail.
Figure 1.
Nucleic acid and amino acid sequence of CiNR1 and SDS/PAGE of in vitro-synthesized CiNR1 protein. (A) The deduced amino acid sequence begins with a presumptive initiation codon at nucleotide 98 but also contains one downstream Met codon at nucleotide 152, which could be an initiator too. Multiple upstream stop codons (underlined in the 5′ region) suggest the first Met codon as the translation start-site. The ORF contains 587 amino acids and is followed by the 3′ untranslated region of 225 bp, which contains a consensus polyadenylation signal (underlined in the 3′ region). Eight cysteines of the DNA binding domain are in bold. (B) SDS/PAGE analysis of l-[35S]-Methionine-labeled protein encoded by the complete cDNA of CiNR1 in rabbit reticulocyte lisate. The translation products were analyzed in 10% SDS polyacrylamide gels. The control line is the translation product of CiNR1 cDNA antisense strand.
Analysis of the protein synthesized by in vitro transcription and translation from the full length coding region of CiNR1 in rabbit reticulocyte lysate revealed that the major protein band migrated ≈67 kDa on SDS/PAGE compatible with the deduced 587-aa sequence (Fig. 1B). Protein from the same lysate failed to show any binding to labeled T3. A second major band of lower molecular weight, present in SDS/PAGE, may be caused by protein synthesized starting from the methionine 19.
Genomic Cloning and Organization.
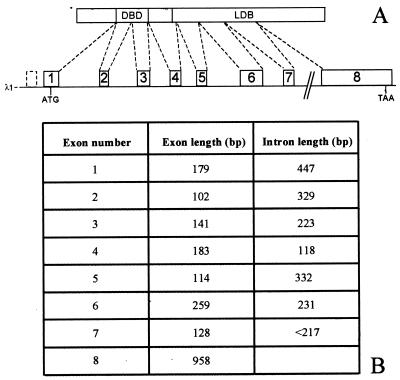
The genomic organization of CiNR1 locus was deduced by sequence analysis of the plasmid insert EcoRI–EcoRI, cloned from λ1, that contains the exons 2–6, the 3′ end of exon 1, and the 5′ end of exon 7 (Fig. 2A). The other exon–intron junctions (exons 7 and 8) were sequenced directly on the phage DNA. This λ1 phage clone appears to contain the whole locus of CiNR1 split in 8 exons distributed on 12 kilobases of genomic DNA (Fig. 2B). The canonical splice consensus sequence is not present in each splicing site (26). The presumptive start methionine is in the middle of exon 1, and exon 8 contains the stop codon.
Figure 2.
Schematic representation of genomic organization of CiNR1 with length of exons and introns. (A) The phagic clone λ1 (12 kilobases long) is composed of 8 exons (white boxes) and 7 introns (lines). The start codon is in the middle of exon 1 and the stop codon is in exon 8. Exons 2 and 3 code for the DBD. The dotted box at the 5′ end of CiNR1 gene is an uncoding exon. Only the 3′ junction of this exon was sequenced. (B) The length of each exon and intron is summarized.
Sequence Comparison and Generation of a Phylogenetic Tree of CiNR1.
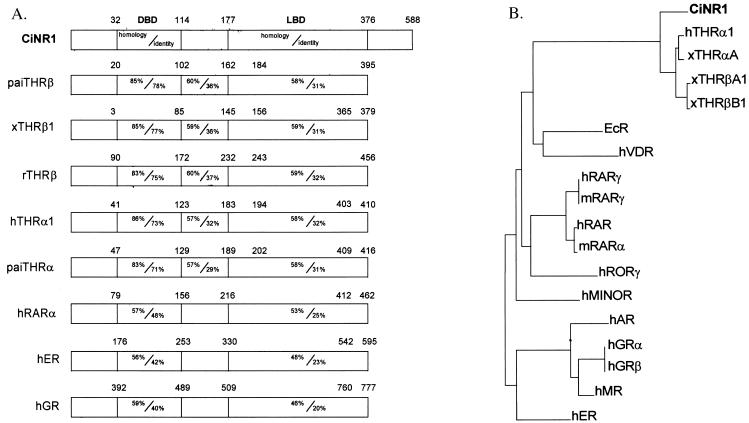
Comparison of the amino acid sequence of CiNR1 with those of other members of the nuclear receptor family revealed the greatest similarity to the TR subfamily (Fig. 3A). The DBD is the more conserved portion and exhibits 86% homology with that of human TRα (27), 85% homology with the DBD of Japanese flounder (Paralichthys olivaceus) TRβ (28) and with that of X. laevis TRβ (15), and 83% homology with rat TRβ (29) and Japanese flounder TRα (30). The P box shows the characteristic sequence EGCKG that belongs to the RAR/TR subfamily (31). In the D domain, the sequences of T box and A box show a good similarity with the same region of other TRs (32).
Figure 3.
Amino acid comparison and evolutionary tree. (A) Primary amino acid sequence comparison of CiNR1 and other members of the nuclear receptor superfamily: paiTRβ (P. olivaceus TRβ, n° D45245); xTRβ (Xenopus laevis TRβ1, n° M35359); rTRβ (rat TRβ, n° J03933); paiTRα (n° D16461); hTRα (human TRα 1, n° M24748); hRARα (n° X06614); hGR (human glucocorticoid receptor, n° X03225); and hER (human estrogen receptor, n° M12674). Amino-acid sequences were aligned by using the Genetics Computer Group program bestfit. Regions of significant similarity between CiNR1 and other receptors are presented schematically as a percent of amino-acid homology/identity. (B) Phylogenetic tree of BDBs of 18 nuclear receptor genes based on amino acid sequences alignment performed by using the growtree phylogram program from the Genetics Computer Group package. We used DBD of hTRα1 (n° M24748), xTRαA (n° M35343), xTRβA1 (n° M35359), xTRβB1 (n° M35361), EcR (ecdisone receptor, n° M74078), hVDR (human vitamin D receptor, n° J03258), hRARγ (n° M24857), mRARγ (mouse RARγ, n° M34476), hRAR (n° X06538), mRARα (n° X57728), hRORγ (n° U16997), hMINOR (n° U12767), hAR (human androgen receptor, n° M20132), hGRα (n° M10901), hGRβ (n° M11050), hMR (human mineral corticoid receptor, n° M16801), and hER (n° X03635).
The ligand binding domain (LBD) of CiNR1 exhibits only 58% homology and 31% identity with the LBD of TRs α and β from Japanese flounder to human. The most terminal carboxy-end part of the Ciona receptor spans ≈200 amino acids more than other TRs. Moreover, the F2 domain of transactivation is not present in CiNR1. The homology of the C-terminal part of CiNR1 with the same region of hTRα2, which shares with CiNR1 a longer LBD than other TRs, is 33%.
Comparison of amino acid sequences of CiNR1 with RARs shows less similarity than that of TRs: an average homology <60% and identity <50% in the DBD and an average homology <55% with identity <25% in the LBD. The homology with other steroid receptors is ≈50% in the DBD and 45% in the LBD. A phylogenetic tree calculated from DBD amino acid sequences places CiNR1 before the duplication of TR into TRα and TRβ (Fig. 3B). Thus, CiNR1 should be considered the ancestor of TR genes.
Expression of CiNR1.
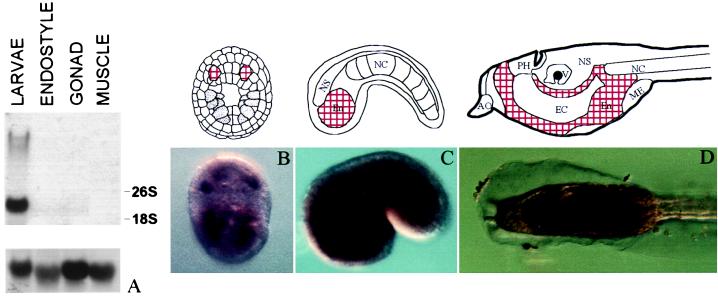
To study the tissue specificity of CiNR1 expression, tissues from whole larvae and adult animals were studied. Northern blot analysis of total RNA from endostyle, gonad, and muscle was performed (Fig. 4A). A 925-bp cDNA fragment encoding the 3′ end of the LBD hybridized to a 2.7 kilobase transcript only in the larval stage whereas adult tissues were virtually negative for CiNR1 expression. The same results were obtained by using the DBD as a probe. To obtain an overall view of CiNR1 localization, in situ hybridization experiments were carried out. No signal was detected at the gastrula stage. Neural plate stage expression of CiNR1 was detected in two regions in the ventral part of the embryo (Fig. 4B) in which the positive cells could belong to the endodermal tissue. At early tailbud stage, a strong signal was observed in the endodermal cells of the head region (Fig. 4C). The same pattern of expression was conserved in the larval stage. The endodermal cells of the tadpole head were intensely positive where the stronger signal localized in the gut primordium and in the cells surrounding the endodermal cavity (Fig. 4D).
Figure 4.
Expression analysis of CiNR1 in adult and embryonic tissues. (A) Northern blot analysis of CiNR1 in different adult and larval tissues. Ribosomal subunits were indicated. After the removal of the first probe, the filter was hybridized with an oligo against a rat ribosomal subunit (Lower) to test the amount of RNA blotted. (B–D) Whole-mount in situ hybridization of CiNR1 in C. intestinalis. (B) Embryo at neural plate stage; vegetal view. (C) Early tail-bud; lateral view. (D) Swimming larva; lateral view. Each stage is joined to a schematic representation. AO, adhesive organ; PH, primordial pharynx; V, vesicle of prosencephalon; NS, nervous system; NC, notochord; En, endoderm; EC, endodermal cavity; ME, mesodermal pocket. Positive regions are found in the endodermal tissues and correspond to the red areas in the schemes.
Transcriptional Activity and DNA Binding of CiNR1.
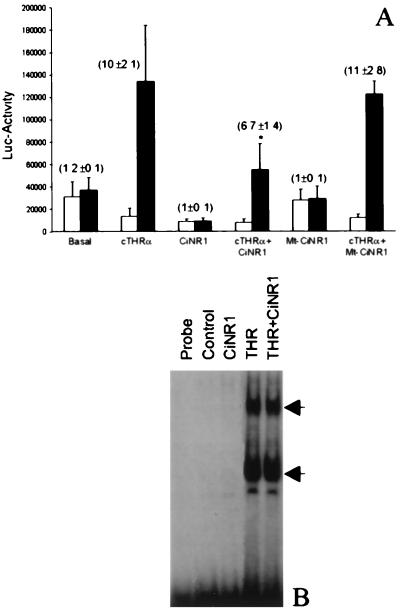
The ability of CiNR1 to induce transactivation was examined by cotransfection experiments by using a TRE-pal driving luciferase as a reporter gene and were compared with cotransfections with the wild-type cTRα. As shown in Fig. 5A, over-expression of CiNR1 reduces the basal activity of this promoter in the presence or absence of T3 treatment whereas T3 increases the transcriptional activity of cTRα-transfected COS7 cells by 10- ± 2.1-fold. When both cTRα and CiNR1 were cotransfected, basal activity of the TRE-pal was reduced to 3.7- ± 1.3-fold. T3 induction was decreased by CiNR1. To investigate which CiNR1 region was responsible of this dominant negative effect, Mt-CiNR1, lacking the C-terminal region, was constructed. The mutated receptor did not affect the basal activity of the promoter and was unable to interfere with cTRα transcriptional activity, suggesting that the C-terminal region of CiNR1 is involved with factors that mediate the induction of transcription. To rule out the possibility that the reduction of promoter activity observed might be caused by competition between CiNR1 and cTRα for binding to DNA, binding of CiNR1 with DNA was studied. In vitro-translated Ciona receptor did not bind PAL oligonucleotide (data not shown), and gel shift assays performed by using extracts of COS7 cells expressing CiNR1 did not exhibit any binding to DNA (Fig. 5B). The same amount of extract from cells transfected with cTRα plasmid showed specific binding to the PAL oligonucleotide and the presence of CiNR1 did not interfere with its DNA binding.
Figure 5.
Transcriptional activity and DNA binding of CiNR1. (A) Dominant negative activity of CiNR1. COS7 cells were transfected with 1 μg of TR, 6 μg of CiNR1, 1 μg of TR, and 6 μg of CiNR1, or 1 μg of TR and 6 μg of Mt-CiNR1 in presence (■) or in absence (□) of 10−7M T3. The luciferase activity was measured in the corresponding cell extracts and was normalized with respect to CAT activity. The figure shows the mean ± SD of three determinations. Values in parentheses are folds of stimulation by T3. P < 0.01 is compared with cTRα alone. (B) DNA binding of TRα and CiNR1. An equal amount (4 μg) of COS7 extracts transfected with empty-plasmid (control), pSG5TRα, or pSG5CiNR1 was used for binding reactions with ≈0.5 ng of 32P-labeled PAL-oligonucleotide. Arrows indicate specific complexes.
DISCUSSION
A member of the nuclear hormone receptor superfamily isolated from the tunicate C. intestinalis, CiNR1, is reported. It contains all of the structural elements of steroid/thyroid hormone receptors: a DBD with two zinc fingers and ligand binding and dimerization domains (1–3). Sequence comparison with known nuclear receptors shows that CiNR1 has the most similarity with TRs. The more conserved domain of CiNR1 is the DBD, which exhibits, at the amino acid level, ≈86% homology with that of other TRs. The P box amino acids are the same as the TR/RAR subfamily. The A box and the T box, regions essential for DNA recognition, are similar in CiNR1 and other TRs (33, 34). In the LBD, the homology with TRs decreases to 58%, and the identity decreases to 31%. The amino acids involved in thyroid hormone binding are not well conserved when comparing the Ciona receptor with other TRs (35), and the C-terminal region of CiNR1 totally differs from other TRs showing, at the C terminus, an extra 200 amino acids. Furthermore, CiNR1 lacks the AF2 transactivation domain. No sequences could be found in GenBank that were homologous to the 200 amino acid terminal sequence of CiNR1. This is reminiscent of TRα2, which has an extra 80 amino acids in the C-terminal region and which does not bind T3 (36). Repeat Northern blots with long exposure failed to demonstrate an alternatively spliced form similar to that found in TRα. Comparison of the CiNR1 terminal region with the same part of TRα2 shows 33% homology. The genomic organization of CiNR1 shows structural similarities to that of TRs; the first exon encodes for the A/B domain, the DBD is encoded by exons 2 and 3, and the last five exons encode the LBD. The position of an intron between the two zinc finger exons is characteristic of the RAR/TR subclass (7), as it is located one amino acid C terminal to the last cysteine of the exon coding for the first zinc finger. Similar to hTRα (37), CiNR1 has a noncoding exon at the 5′ untranslated region. Northern blot analysis confirms that CiNR1 mRNA spans ≈700 nucleotides 5′ to the cDNA clone we sequenced. Phylogenetic analyses show that CiNR1 is likely to be the ancestor gene of the TRs.
Dominant negative activity was observed when CiNR1 was cotransfected with cTRα in COS7 cells, reminiscent of the same activity seen with the α2 form of TR, and, in both cases, the dominant negative activity occurs without DNA binding (38). The formation of inactive heterodimers or the depletion of accessory factors are possible explanations. The ability of CiNR1 to repress basal transcription in absence of ligand is in favor of this second hypothesis. The lack of these activities in Mt-CiNR1 places the important factor in the C-terminal part of this protein. Unfortunately, because this region is essential both for dimerization and for interaction with general transcription factors, it cannot discriminate between these two mechanisms. The functional role of CiNR1 is under investigation, but its spatiotemporal expression provides some clues at present. Indeed, by Northern blot assay, we showed that the CiNR1 is expressed only in the larval stage, being absent in all adult tissues tested. Furthermore, in situ hybridization showed that the expression of CiNR1 is specific for the endodermal tissue exclusively from neural plate to larval stage, suggesting that CiNR1 may trigger a specific nuclear signal involved in the development and metamorphosis of vertebrate progenitors. Finally, we are left with the enigma that, in one of the most primitive species to synthesize thyroid hormone, which then is involved in its metamorphosis, we find a nuclear receptor that by sequence is closely related to known thyroid hormone receptors and yet cannot bind thyroid hormone.
Acknowledgments
We are grateful to Dr. H. G. Stunnenberg for 3TRE-Pal and PSG5cTHRα plasmids. We also thank Prof. M. D’Armiento and Dr. S. Dolci for helpful comments on the manuscript. We are indebted to the late Prof. Gaetano Salvatore for his continued advice and encouragement. We are also in debt to Dr. Zdeneck Kostrouch who, with his science, suggestions, and friendship, made possible this work. This work was supported by the Target Project on Biotecnology and by Italian Ministero dell’Universitá e della Ricerca Scientifica e Technologica and Consiglio Nazionale delle Ricerche grants.
ABBREVIATION
- CiNR1
Ciona intestinalis nuclear receptor 1
- TR
thyroid receptor
- RAR
retinoic acid receptor
- DBD
DNA binding domain
- LBD
ligand binding domain
- CAT
chloramphenicol acetyl transferase type I
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF077403).
References
- 1.Yamamoto K R. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- 2.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai M J, O’Malley B W. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 4.Enmark E, Gustafsson J A. Mol Endocrinol. 1996;10:1293–1307. doi: 10.1210/mend.10.11.8923456. [DOI] [PubMed] [Google Scholar]
- 5.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesomo K, Blumberg B, Kastener P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laudet V. J Mol Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- 7.Laudet V, Hänni C, Coll J, Catzeflis F, Stéhelin D. EMBO J. 1992;11:1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amero S A, Kretsinger R H, Moncrief N D, Yamamoto K R, Pearson W R. Mol Endocrinol. 1992;6:3–7. doi: 10.1210/mend.6.1.1738368. [DOI] [PubMed] [Google Scholar]
- 9.Patricolo E, Ortolani G, Cascio A. Cell Tissue Res. 1981;214:289–301. doi: 10.1007/BF00249213. [DOI] [PubMed] [Google Scholar]
- 10.Roche J, Salvatore G, Rametta G. Biochim Biophys Acta. 1962;63:154–165. doi: 10.1016/0006-3002(62)90348-7. [DOI] [PubMed] [Google Scholar]
- 11.Denucé J M. Z Naturforsch. 1991;46:1094–1100. doi: 10.1515/znc-1991-11-1227. [DOI] [PubMed] [Google Scholar]
- 12.Katz M J. Biol Bull. 1983;164:1–27. [Google Scholar]
- 13.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S, Degnan B M, Ross I L, Hawkins C J, Lavin M F. Mar Biol (Berlin) 1988;98:95–100. [Google Scholar]
- 15.Yaoita Y, Shi Y B, Brown D D. Proc Natl Acad Sci USA. 1990;87:7090–7094. doi: 10.1073/pnas.87.18.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manfioletti G, Schneider C. Nucleic Acids Res. 1988;16:2873–2884. doi: 10.1093/nar/16.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swofford D L, Olsen G J. In: Molecular Systematics. Hillis D M, Moritz C, editors. Sunderland, MA: Sinauer; 1990. pp. 411–500. [Google Scholar]
- 20.Caracciolo A, Gesualdo I, Branno M, Aniello F, Di Lauro R, Palumbo A. Dev Growth Differ. 1997;39:437–444. doi: 10.1046/j.1440-169x.1997.t01-3-00004.x. [DOI] [PubMed] [Google Scholar]
- 21.Sap J, Munoz A, Dumm K, Goldberg Y, Ghysdal J, Vennstrom B. Nature (London) 1986;324:635–640. doi: 10.1038/324635a0. [DOI] [PubMed] [Google Scholar]
- 22.Samuels H H, Stanley F, Casanova J. Endocrinology. 1979;105:80–85. doi: 10.1210/endo-105-1-80. [DOI] [PubMed] [Google Scholar]
- 23.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umesono K, Evans R M. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 25.Luisi B F, Xu W X, Otwinowski Z, Freedman L P, Yamamoto K R, Sigler P B. Nature (London) 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 26.Breathnach R, Chambon P. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- 27.Weinberger C, Thompson C C, Ong E S, Lebo R, Gruol D J, Evans R E. Nature (London) 1986;324:641–646. doi: 10.1038/324641a0. [DOI] [PubMed] [Google Scholar]
- 28.Yamano K, Inui Y. Gen Comp Endocrinol. 1995;99:197–203. doi: 10.1006/gcen.1995.1102. [DOI] [PubMed] [Google Scholar]
- 29.Thompson C C, Weinberger C, Lebo R, Evans R M. Science. 1987;237:1610–1614. doi: 10.1126/science.3629259. [DOI] [PubMed] [Google Scholar]
- 30.Yamano K, Araki K, Sekikawa K, Inui Y. Dev Genet (Amsterdam) 1994;15:378–382. doi: 10.1002/dvg.1020150409. [DOI] [PubMed] [Google Scholar]
- 31.Forman B M, Samuels H H. Mol Endocrinol. 1990;4:1293–1301. doi: 10.1210/mend-4-9-1293. [DOI] [PubMed] [Google Scholar]
- 32.Glass C K. Endocr Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 33.Wilson T E, Paulsen R E, Padgett K A, Milbrandt J. Science. 1992;256:107–110. doi: 10.1126/science.1314418. [DOI] [PubMed] [Google Scholar]
- 34.Lee M S, Kliewer S A, Provencal J, Wright P E, Evans R M. Science. 1993;260:1117–1121. doi: 10.1126/science.8388124. [DOI] [PubMed] [Google Scholar]
- 35.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. Nature (London) 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 36.Mitsuhashi T, Tennyson G E, Nikodem V. Proc Natl Acad Sci USA. 1988;85:5804–5808. doi: 10.1073/pnas.85.16.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laudet V, Bengue A, Henry-Duthoit C, Joubel A, Martin P, Stéhelin D, Saule S. Nucleic Acids Res. 1991;19:1105–1112. doi: 10.1093/nar/19.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu R T, Suzuki S, Miyamoto T, Takeda T, Orata M, De Groot L J. Mol Endocrinol. 1995;9:86–95. doi: 10.1210/mend.9.1.7760853. [DOI] [PubMed] [Google Scholar]