Abstract
The tobacco mosaic virus (TMV) movement protein (MP) facilitates transport of virus infection between adjacent cells by modifying plasmodesmata. Previous studies suggested that the cytoskeleton and the endomembrane system are involved in this transport. We examined the effects of TMV infection on the endoplasmic reticulum (ER) in transgenic Nicotiana benthamiana that accumulate the green fluorescent protein (GFP) in the ER. Fluorescence microscopy was used to show that early in infection the ER undergoes dramatic morphological changes that include the conversion of tubular ER into large aggregates that revert to tubular ER in later stages of infection. These changes parallel MP accumulation and degradation. Furthermore, a fusion protein comprising MP fused to GFP accumulates in or on these large aggregates of ER. Expression of MP-GFP in the absence of virus infection led to the production of fluorescent aggregates of the same apparent form and size. Microsomes isolated from infected leaves contain MP. We show that the MP appears to behave as an integral ER membrane protein and is exposed on the cytosolic face of the ER. The importance of the association of MP with ER and its possible role in intracellular and intercellular spread of infection is discussed.
Viruses establish a successful infection in plants by exploiting cytoplasmic bridges, the plasmodesmata (Pd), that connect neighboring cells (1, 2). Pd form a channel which is spanned by plasma membrane and contains a thin, appressed tubule of the endoplasmic reticulum (ER) (desmotubule) thereby establishing cytoplasmic as well as ER continuity between neighboring cells (3). Under most conditions Pd allow the transport of relatively small molecules (<1 kDa), and to move to adjacent cells viruses must alter Pd to allow transport of larger molecules. Many viruses express proteins that are involved in cell–cell spread, referred to as movement proteins. Tobacco mosaic virus (TMV) forms rod-shaped particles and has a single-stranded, plus sense RNA genome that encodes proteins involved in genome amplification (replicase), the 30-kDa movement protein (MP) and the 17.5-kDa coat protein (CP). Whereas the replicase and CP RNAs are produced continuously during infection, the MP RNA is produced transiently (4).
The TMV MP is essential and sufficient for local cell–cell spread of the infection (5, 6), whereas in most host plants long-distance spread also requires CP (7). Electron microscopy and fluorescence microscopy studies localized MP (8, 9) and MP-GFP [fusion between MP and green fluorescent protein (GFP)] produced during viral infection (10) and in transgenic plants to Pd. MP modifies the size exclusion limit of Pd (11, 12) as indicated by dye-coupling studies and cooperatively binds to single-stranded nucleic acid in vitro (13, 14). When isolated from virus-infected tissue or transgenic plants, the MP is associated with a cell wall fraction as well as with membrane fractions (5, 15–17). Studies of MP-GFP fusion proteins showed that in infected protoplasts the protein coaligns with microtubules and microfilaments (18, 19). On the basis of these studies it was suggested that the cytoskeleton is involved in the transport of MP and ribonucleoprotein complexes that contain MP, to the Pd and across the cell wall.
It was previously shown that the cellular endomembrane system is involved in establishing infection by TMV (20–23) and by other plant viruses (24–27). In a recent study, fluorescence microscopy was used to investigate colocalization of MP-GFP with ER in infected tobacco protoplasts and to follow its accumulation in “irregular shaped structures” that were suggested to be derived from ER (28).
Here we investigate the nature of the association of MP with ER during TMV infection. Fluorescence microscopy of infection sites on Nicotiana benthamiana that produce an ER-targeted GFP (erGFP) revealed a tight association of MP (and MP-GFP) with ER. Furthermore, we observed severe morphological changes of the ER during infection that are followed by recovery to the wild-type appearance; recovery is coincident with removal of MP-GFP. The aggregates of MP and ER that are formed during virus infection are also observed when MP-GFP is expressed in the absence of virus infection. Biochemical analyses of isolated microsomes confirm a tight association of MP with ER and suggest that MP is exposed on the cytoplasmic face of the ER and is most probably an integral membrane protein. A model for cell–cell spread of TMV is proposed that involves transport of the MP along the ER toward the Pd and across the cell wall.
MATERIALS AND METHODS
Constructs and Plants.
Infectious transcripts were prepared from linearized plasmids pTMV-M:Gfus (18) (subsequently referred to as pTMV-MP:GFP), pU3/12–4 (29), and pTMV-ΔC-MP*. pTMV-ΔC-MP* encodes MP with six histidine residues added at the C-terminus and lacks the CP gene. It was constructed by BsmBI–NotI digestion of pTMV-MP:GFP, Klenow DNA polymerase treatment of the NotI site to create a blunt end, and insertion of a BsmBI–SmaI fragment from pGEX-MPhis (T. Hu and R.N.B., unpublished work), carrying the MP-His6. TMV-ΔC-MP* was equally infectious (i.e., it was capable of local spread) as TMV and TMV-MP:GFP. Plasmid pe35S-MP:GFP contains an enhanced cauliflower mosaic virus (CaMV) 35S promoter (30) and the full-length MP fused to GFP. MP was amplified by PCR to introduce the 5′ BamHI restriction site (5′ oligo: GCGGGATCCGCGGCCGCACATATGGCTCTAGTTGTTAAAGG) and the 3′ EcoRI restriction site (3′ oligo: CCGCGGCCGCGAATTCTATTTAAAACGAATCCGATTCGGCGACAGTAGCC) and was cloned as a BamHI–EcoRI fragment into pUC19. Site-directed mutagenesis was used to delete the stop codon and introduce an EcoRV restriction site (oligo: CGGATTCGTTTGGGATATCGAATTCACTGGCC). A SmaI–BamHI fragment from a pTMV-MP:GFP analog (pTMV-MP:GFP[M153T,V163A]), containing GFP with amino acid changes S65T, M153T, and V163A to increase fluorescence, was inserted along with the BamHI–EcoRV fragment from the above plasmid into the BamHI site of pUC19. Transfer of a BamHI fragment into pMON921 after BglII and BamHI digestion, positioning the MP-GFP fusion under the control of the CaMV 35S promoter, resulted in pe35S-MP:GFP.
Nicotiana benthamiana plants that accumulate ER-targeted GFP (erGFP) contain a GFP expression cassette constructed by Haseloff and colleagues (31).
Fluorescence Microscopy.
The fluorescence microscopy was performed as described by Kahn et al. (32). Leaf disks containing infection sites were excised and mounted as live tissue on microscope slides and investigated immediately.
Particle Bombardment.
Particle bombardment was carried out with a helium-driven gene gun (Biolistic PDS-1000/He, Bio-Rad) with 1100-psi rupture disks. Coating of DNA was performed on 1-μm gold beads. Leaves were incubated for 15–20 hr after bombardment at room temperature in the dark.
Preparation of Microsomal Fractions.
Microsome-containing fractions were prepared from inoculated leaves (2 g) of N. benthamiana erGFP plants by homogenization in buffer A [250 mM Tris⋅HCl, pH 8/10 mM KCl/1 mM EDTA/0.1 mM MgCl2 or 5 mM MgCl2/0.1% BSA/10% (wt/vol) sucrose/1 mM DTT/1 mM phenylmethanesulfonyl fluoride (PMSF)/10 μM E-64 and 50 μM MG115 (protease inhibitors, Peptides International)/1 μM pepstatin/10 μg/ml ALLM (protease inhibitor, Sigma)], and the homogenate was filtered through Miracloth (Calbiochem). After centrifugation at 3,800 × g, the supernatant was centrifuged at 100,000 × gav for 45 min to collect the membrane fraction. This fraction was resuspended in buffer A and loaded onto 18–55% (wt/vol) sucrose gradients in buffer B (50 mM Tris⋅HCl, pH 8/10 mM KCl/1 mM EDTA/0.1 mM MgCl2 or 5 mM MgCl2/0.1% BSA). Gradients were centrifuged 8 hr at 100,000 × gav and 1-ml fractions were collected. Aliquots were diluted in loading buffer and 10 μl of each fraction was subjected to Western blot analyses.
Proteinase K Treatment.
Membranes from sucrose gradient fractions were collected by centrifugation at 140,000 × gav for 45 min and resuspended in buffer B containing 10% (wt/vol) sucrose and 0.1 mM MgCl2 (buffer C). Aliquots were subjected to proteinase K digestion (0.1 g/liter) for 20 min on ice in the absence or presence of Triton X-100 (0.4%). Samples were collected by adding 2× SDS loading buffer containing 4 mM EGTA, boiled 5 min, and frozen for later analysis. To assay for endogenous protease activity aliquots were incubated in the absence of proteinase K.
Chemical Treatment of Membrane Fractions.
Membranes collected from sucrose gradient fractions were resuspended in buffer C. Aliquots were incubated 30 min on ice with 2.5 M urea, 0.5 M NaCl, 1% Triton X-100, or 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) (in buffer C). Samples were then subjected to centrifugation at 100,000 × gav for 45 min; pellets were rinsed once with buffer C and resuspended in loading buffer.
Western Blot Analysis.
Nitrocellulose filters were blocked with 2% (wt/vol) nonfat milk powder and incubated in primary antibody overnight; anti-MP antibody was raised against residues 209–222 of the MP (32). The polyclonal anti-GFP antibody was from CLONTECH, anti-BiP (immunoglobulin light chain binding protein) antibody was a gift from R. Boston (North Carolina State University, Raleigh), and the anti-H+-ATPase antibody was a gift from B. Hong and J. Harper (33). Incubation with secondary antibody (anti-rabbit IgG, conjugated to horseradish peroxidase) was for 90 min. Blots were developed with substrate for chemiluminescence (SuperSignal ULTRA, Pierce).
RESULTS
Fluorescence Microscopy.
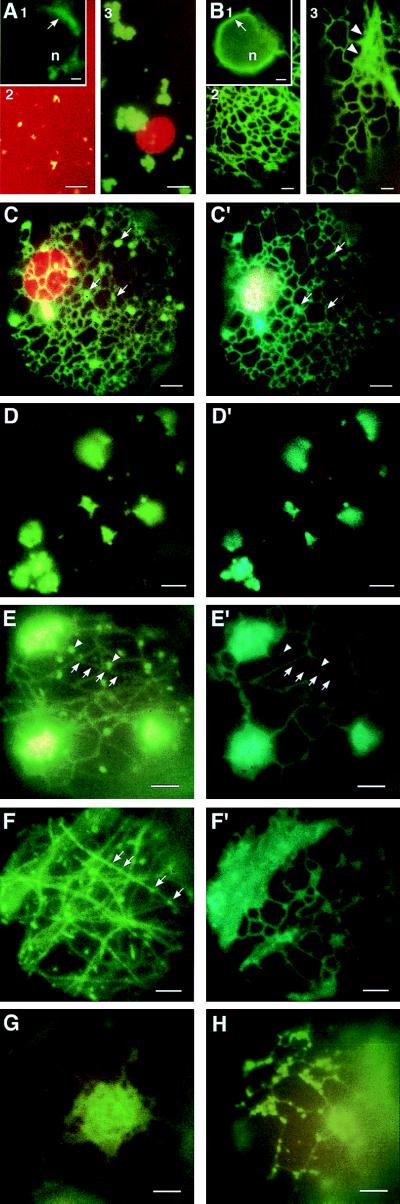
To study the impact of TMV infection on plant ER, live cell imaging by conventional fluorescence microscopy was used to observe inoculated leaves of wild-type N. benthamiana and of transgenic N. benthamiana that accumulate erGFP. Nontransgenic N. benthamiana plants infected by TMV that was modified to produce MP-GFP fusion protein (TMV-MP:GFP) develop fluorescent, expanding ring-shaped infection sites (34, 35). Detailed investigation of the outermost cells of these infection sites reveals very small, rapidly moving, granular fluorescent particles at the cell cortex (Fig. 1A2). Fluorescent material is also detected as a halo around the nucleus (Fig. 1A1, arrow) and in fast-flowing streams that traverse the cytoplasm (not shown), whereas in adjacent cells, toward the center of the ring, most of the fluorescence was associated with larger aggregates (Fig. 1A3). Under these conditions there was no green fluorescence from noninfected tissue detectable (not shown).
Figure 1.
Fluorescence micrographs of epidermal leaf cells of N. benthamiana (A) or N. benthamiana erGFP (B–H) infected with TMV or TMV-MP:GFP. (A1) Fluorescence surrounding the nucleus (n) due to MP-GFP early in infection (arrow). (A2) Fluorescent structures containing MP-GFP in cells early in infection. (A3) Cortical fluorescent bodies in early to mid stages of infection. (B) Noninfected epidermal cells of N. benthamiana erGFP. (B1) Perinuclear fluorescent halo of erGFP (arrow). (B2) Normal reticulate pattern of cortical ER. (B3) Fast-moving tracks of erGFP fluorescence (arrowheads). (C–F) Left panels excited with blue light, right panels excited with UV. Note: UV excitation exclusively excites erGFP, while excitation with blue light excites both MP-GFP and erGFP. (C, C′) Early stage of infection, showing small cortical aggregates formed at vortexes of ER network; much of the fluorescence is contributed by MP-GFP (arrows). (D, D′) Large cortical aggregates and disrupted cortical ER in mid-stages of infection. (E, E′) Recovery of tubular ER and dissociation of large cortical aggregates and development of filamentous fluorescence of MP-GFP colocalized with microtubules (arrows) as well as a punctate fluorescence on the cell surface (arrowheads) in mid to late stages of infection. (F, F′) Cortical aggregates decrease and filaments comprising MP-GFP on microtubules (arrows) are apparent in late stages of infection; punctate fluorescent structures often aligned with microtubules (arrows); ER has recovered to preinfection state. (G) Large cortical ER aggregates induced in N. benthamiana erGFP during infection by wild-type TMV. (H) Tubular cisternae of the cortical ER are disrupted and converted to lamellar aggregates. (Bars = 0.5 μm.)
These observations, coupled with a previous study suggesting that in mid-to-late stages of infection MP-GFP is associated with ER (28), led us to investigate the role of ER in cell–cell movement of virus infection. For these studies we selected a transgenic line of N. benthamiana that accumulates GFP in the ER (erGFP) as a host for TMV. In these plants the ER was highly fluorescent and certain regions, including the perinuclear area, were in constant motion that presumably represents movement of the ER (Fig. 1B1, arrow, and Fig. 1B3, arrowheads). The motion of the ER was similar to the motion of fluorescent particles described in plants infected with TMV-MP:GFP (Fig. 1A). In addition, the reticulate network of tubular ER elements and intervening lamellar cisternae comprising the cortical ER was readily visible in these tissues (Fig. 1 B2 and B3). The tubular elements are in constant motion around fixed sites, changing their position, often loosing their connections to lamellar cisternae and establishing new connections to the vortexes of the ER. These characteristics have previously been reported for ER in plant cells (36–38).
Transgenic plants that contain erGFP were infected with TMV-MP:GFP, and the ring-shaped, fluorescent infection sites that formed were examined by fluorescence microscopy between 3 and 5 days after infection. Cells within these expanding fluorescent rings reflect an approximate time course of the virus infection cycle (18). Fluorescence emitted by MP-GFP can be distinguished from fluorescence of erGFP because the excitation spectra of the two GFP variants are different (31, 39). UV excites erGFP alone [excitation maximum (exmax) = 395 nm], whereas blue light excites both erGFP (exmax = 473 nm) and MP-GFP (exmax = 488 nm). By comparing the pattern of fluorescence after UV excitation with that observed after excitation by blue light, we distinguished between ER labeled by erGFP and fluorescent structures that contain MP-GFP. In some of the micrographs a very low degree of “bleed-through” from the blue-light channel into the UV channel was observed (in Fig. 1, compare E with E′, F with F′), but it was not significant for the interpretation of the results.
At the leading edge of the infection rings, fluorescent aggregates of MP-GFP were observed on the vortexes of the cortical ER; these aggregates grow in size during infection (Fig. 1C; compare with 1A3). When these tissues were illuminated with UV (Fig. 1C′), and compared with fluorescence after blue-light excitation (Fig. 1C) it is apparent that most of the fluorescence in these aggregates is derived from MP-GFP (arrows).
In later stages of infection (moving inwards in the ring, toward the center of the infection site) a more dramatic change in the morphology of the ER is observed. Most of the infected cells contain a few large fluorescent cortical bodies that continue to grow in size and are excited by both UV and blue light (Fig. 1 D, D′, E, E′). The strands of ER that interconnect these bodies either are disrupted or are stretched to form long thin “lines” (Fig. 1 D and D′); most of the vivid vibrations and rearrangements of the ER tubules that are observed in noninfected cells (as described above) are not evident in these cells. There was clear colocalization of MP-GFP (Fig. 1 D and E) and what appears to be aggregates of the ER that contain erGFP (Fig. 1 D′ and E′), indicating a close association of MP-GFP and ER. In neighboring cells and cells closer to the center of the infection ring—i.e., cells that are in later stages of the infection cycle—the ER recovers to the appearance of ER in noninfected cells (Fig. 1F′) and MP-GFP is associated with filaments (Fig. 1 E and F, arrows), previously shown to be microtubules that colocalize with MP-GFP (18, 19). Punctate fluorescent bodies are observed at the surface of the epidermal cells (Fig. 1E, arrowheads, and F, arrows). In some cases these fluorescent points clearly align with microtubules and are not fluorescent under UV excitation (Fig. 1F, arrows). We conclude that these structures consist mainly of MP-GFP rather than ER.
Throughout the diameter of the fluorescent ring and in the center of the infection site MP-GFP is also associated with punctate sites in the cell wall (not shown); these sites were recently shown to represent Pd that contain MP-GFP (10).
When N. benthamiana erGFP plants were infected by nonfluorescent wild-type TMV, the infection induced significant changes in the ER (Fig. 1 G and H) that are similar to those described above (Fig. 1 C–F). The ER appeared to aggregate to form large, mainly cortical bodies (Fig. 1G), and the interconnecting tubular ER cisternae were disrupted or became increasingly thinner (Fig. 1H); in adjoining cells that were in later stages of infection the ER was restored to the normal structures (not shown). These results indicated that the changes in ER are reflective of a biological function of the TMV MP per se rather than a nonspecific effect induced by the MP-GFP fusion protein.
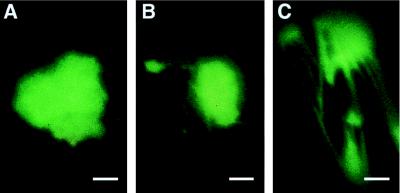
We used particle bombardment of plasmid DNA encoding MP-GFP expressed from the cauliflower mosaic virus 35S promoter to study MP localization in the absence of virus infection. In leaves bombarded with this plasmid, MP-GFP accumulated in fluorescent bodies that resembled in form, size, and location (Fig. 2 A–C) the bodies that are induced during virus infection. In some cells these aggregates were associated with fluorescent, filamentous structures (Fig. 2 B and C). In contrast, cells that produced free GFP showed diffuse fluorescence (not shown). Thus, MP-GFP induced the formation of aggregates that resemble the MP-ER aggregates observed during virus infection (Fig. 1 C–E). MP-GFP fluorescence was also observed in a punctate pattern associated with cell walls (not shown), presumably being sites of Pd. Therefore, MP-GFP expressed in the absence of virus infection appeared to accumulate in all sites to which MP-GFP localizes during virus infection.
Figure 2.
Fluorescence micrographs of N. benthamiana leaves after particle bombardment with pe35S-MP:GFP. Detached leaves were investigated by fluorescence microscopy 15–20 hr after bombardment. In epidermal cells MP-GFP localized to large fluorescent aggregates (A), which often were associated with filamentous structures (B and C) and presumably correspond to MP-GFP coalignment with microtubules. (Bars = 0.5 μm.)
Biochemical Characterization of Microsomal ER.
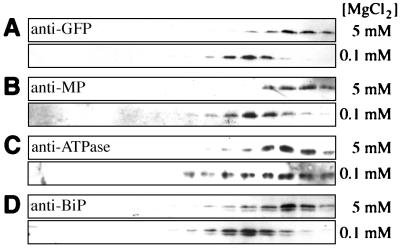
Leaf tissue infected with a mutant of TMV that produces MP that contains 6 histidine residues at the C terminus (TMV-ΔC-MP*) was used to analyze the nature of the association of the MP with ER. Two days after infection, microsomal fractions were prepared from inoculated leaves either in the presence of 5 mM MgCl2 to preserve ribosome binding or in the presence of 0.1 mM MgCl2 to release the ribosomes. Membranes were subjected to sucrose gradient fractionation and Western blot analyses of the gradient fractions. Immunodetection of erGFP and BiP showed that, as expected, erGFP and BiP accumulate in fractions of high mobility in these gradients (e.g., high density); membranes treated with 0.1 mM MgCl2 displaced erGFP and BiP by 3 fractions compared with those prepared in the presence of 5 mM MgCl2 (Fig. 3, A and D). These results confirm that erGFP (31) and BiP are associated with ER and that erGFP can be used as ER marker in these experiments. As a control we used an antibody specific for a proton-ATPase that is known to reside exclusively in the plasmalemma (33). The position of the ATPase in the gradient was not significantly affected by treatment with 0.1 mM MgCl2 (Fig. 3C).
Figure 3.
Western blot analyses of fractions from sucrose density gradients. Membrane preparations with bound (5 mM MgCl2) or displaced (0.1 mM MgCl2) ribosomes were separated on an 18–55% sucrose gradient. Fractions were analyzed with indicated antibodies. Sedimentation was from left to right. (A) Anti-GFP. (B) Anti-MP. (C) Anti-ATPase. (D) Anti-BiP.
When gradient fractions were analyzed with anti-MP antibody, a distribution similar to that observed for erGFP was detected—i.e., the fractions containing MP were also shifted by the treatment with 0.1 mM MgCl2, suggesting that MP associates with ER in planta (Fig. 3B). Similar results were obtained with leaf tissue from plants infected by wild-type TMV (not shown).
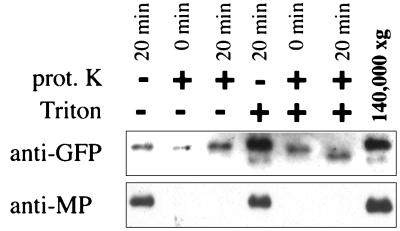
To investigate the nature of the association of MP with ER in greater detail, microsome fractions that contain MP were collected and treated with proteinase K in the absence or presence of Triton X-100. As expected for a luminal ER protein, erGFP was resistant to proteinase K digestion in the absence of detergent (Fig. 4). However, in the presence of detergent there was a clear decrease in size of GFP, indicating that the protein was exposed to proteinase K. It has been previously reported that GFP is highly resistant to protease digestion (40), which likely is a result of its barrel-shaped three-dimensional structure (41, 42), and explains why erGFP was not completely degraded by proteinase K in the presence of detergent. In contrast, MP was susceptible to proteinase K in the presence or absence of detergent (Fig. 4). This observation indicates that at least a C-terminal portion of the MP is exposed to the cytosolic face of the isolated microsomes, because the antibody used in this study was raised against a C-terminal peptide of the MP.
Figure 4.
Proteinase K treatment of fractions from sucrose gradients. Microsomal membranes were collected from fractions of the sucrose gradients (see Fig. 3) and subjected to proteinase K digestion in the absence (−) or presence (+) of Triton X-100 and analyzed by Western blotting with anti-GFP (Upper) or anti-MP (Lower) antibodies.
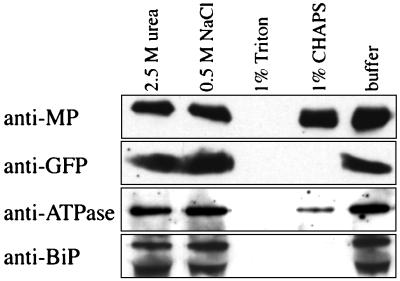
To further characterize the MP-ER association, microsomal preparations were treated with agents that are reported to remove peripherally associated proteins (2.5 M urea; 0.5 M NaCl; 0.1 M Na2CO3) or to disrupt membranes and solubilize integral membrane proteins (1% Triton X-100; 1% CHAPS). After treatment the membranes were collected by centrifugation and investigated by western blot analysis. The association of erGFP and BiP with microsomes was not affected by treatment with agents known to remove peripherally bound proteins, and was clearly disrupted upon treatment with both detergents (Fig. 5), as was expected for ER luminal proteins. The ATPase, which is an integral plasma membrane protein, was dissociated from the microsomes by treatment with Triton X-100 and partially dissociated upon incubation with CHAPS (Fig. 5). While treatment with urea, NaCl, or CHAPS (Fig. 5) or with Na2CO3 (data not shown; ref. 17) did not affect the association of MP with membranes, treatment with the nonionic detergent Triton X-100 disrupted the MP–ER association (Fig. 5). On the basis of these experiments we conclude that MP behaves as an integral membrane protein.
Figure 5.
Effects of membrane washes on association of MP with ER. Microsomal membranes were collected as in Fig. 4 and incubated 30 min on ice with 2.5 M urea, 0.5 M NaCl, 1% Triton X-100, or 1% CHAPS. After samples were collected by centrifugation, they were subjected to Western blot analysis with anti-MP, anti-GFP, anti-ATPase, and anti-BiP antibodies.
DISCUSSION
The TMV MP plays a pivotal role in cell–cell spread of infection; however, the association of MP with cellular components, especially the endomembrane system, has not been studied in detail. When Deom et al. (16) studied the localization of MP by crude fractionation into cell wall fraction, P30 (30,000 × g pellet), and S30 fractions (“soluble” fraction) up to 120 hr after infection, they observed high levels of MP in the P30 and S30 fractions in early stages of infection. These later decreased compared with MP accumulating in the cell wall fraction, where it remained at high levels. These observations suggest that in early stages of infection a considerable proportion of the MP is associated with membranous material rather than with the cell wall fraction.
In our studies fluorescence microscopy of live tissue from leaves from N. benthamiana in early stages of infection by TMV-MP:GFP revealed small fluorescent aggregates of MP-GFP that were not described in earlier studies (10, 18, 19, 34) and exhibited only a dim fluorescence. This particulate fluorescence was often seen on the cell cortex (Fig. 1A2) as well as surrounding the nucleus (Fig. 1A1). The distribution of these aggregates and their movement along “invisible” tracks in the cytoplasm and our observation of a similar type of highly mobile fluorescence in noninfected leaves of N. benthamiana that produce erGFP (Fig. 1B) is reminiscent of the distribution of ER in plants and with the description of organelle movement along ER (3, 43).
In onion bulb epidermal cells ER can be divided into two major subclasses: (i) stationary cortical ER that consists of a network of tubular and lamellar cisternae, and (ii) fast-flowing tubular strands that traverse vacuoles and spread from the perinuclear region toward the cortex (3, 37, 43). Our observations support the suggestion that early after infection the mobile, fluorescent MP-GFP is associated with ER and is transported throughout the cell along the tracks of ER; we suggest that these bodies later aggregate to form the cortical, fluorescent bodies (Fig. 1A3).
Fluorescence microscopy of N. benthamiana expressing erGFP revealed a change in the morphology of the ER upon infection by wild-type TMV. The change in ER resembles the changes (i.e., disruption) of ER in tissue systemically infected by tobacco etch potyvirus (26). On leaves inoculated with TMV, tubular components of the cortical ER lost their dynamic appearance and became increasingly thinner; in addition enlarged aggregates of lamellar cisternae were formed at the vortexes of the ER network (Fig. 1 G and H). These aggregates of ER resembled in size and form the cortical bodies of MP-GFP that were observed upon infection by TMV-MP:GFP on N. benthamiana (Fig. 1A3) (18).
Infection of N. benthamiana erGFP with TMV-MP:GFP allowed us to distinguish GFP produced by the host and the virus, and it made possible an in vivo investigation of the intracellular localization of MP-GFP more detailed than earlier studies that used a single GFP (10, 18, 19, 34, 35). As a result of infection by TMV-MP:GFP, fluorescent aggregates were detected that formed on the vortexes of the cortical ER (Fig. 1 C–E).
Interestingly, MP-GFP alone (i.e., in the absence of virus infection) induced the formation of MP aggregates, which appear to be structurally related to the aggregates of MP and ER observed during virus infection (Fig. 2 A–C). On the basis of these observations and the tight association of MP and microsomal fractions (Fig. 3), we conclude that these structures contain aggregated, lamellar ER associated with MP-GFP.
It has been suggested, on the basis of correlative electron microscopy studies, that the lamellar cisternae of the cortical ER contain rough ER, whereas the tubular structures consist of smooth ER (3). Furthermore, it has been proposed that the default conformation of the ER membrane is tubular, while the “physical constraints” of ribosome binding convert tubules into lamellar cisternae (3). It is tempting to suggest that during TMV infection the invading virus induces the transition from smooth to rough ER (see Fig. 1) by recruiting the binding of ribosomes to ER, to establish the necessary infrastructure for protein synthesis and virus replication. It was previously shown that TMV replication occurs on membranous structures (20–22). Replication of other plant RNA viruses has also been described as taking place in close association with membranes (24–27). Our observations suggest that MP has the intrinsic property to induce ER aggregation to form these structures.
In mid-to-late stages of infection, fluorescent filaments that contain MP-GFP are observed; similar structures have been described in tobacco protoplasts and have been suggested to be MP-GFP that is coaligned with microtubules (18, 19). In cells containing filamentous structures the ER recovers and regains the morphology and mobility that is common in noninfected tissue (Fig. 1 E and F). We suggest that the recovery of the ER corresponds to the release of bound ribosomes, thereby releasing aggregated ER; this presumably coincides with the end of a phase of virus replication that requires the membrane aggregates. We propose that the abrupt disappearance of MP in late stages of the infection cycle indicates the involvement of a targeted degradation of the MP and may be important to allow the ER to regain the “preinfection” conformation.
In the center of the infection site, where the ER is indistinguishable from ER in noninfected tissue, MP-GFP is detected in cell wall-associated points that were identified as Pd (10, 18). Other punctate fluorescent structures are located on the upper surface of the epidermal cells (Fig. 1 E and F). These structures may correspond to structures described in protoplasts infected with TMV-MP:GFP as peripheral punctate fluorescence; it was proposed that these structures are sites that anchor cytoskeleton to membrane and cell wall (28). However, in our study these sites, some of which are aligned with microtubules (Fig. 1 E and F), are not closely associated with ER (Fig. 1E, arrowheads), as in infected protoplasts (28). Therefore, we cannot conclude that punctate structures in epidermal cells and protoplasts are equivalent.
To further characterize the association of the MP with ER, we conducted biochemical analyses of microsome fractions in which MP copurified with the ER markers erGFP and BiP (Fig. 3 A, B, and D), confirming the results of the fluorescence microscopy. Previous cell fractionation studies with tissue from plants that accumulate MP as well as from plants infected by TMV localized MP to cell wall fractions and membrane fractions containing plasmalemma and ER (5, 15–17). Our observations demonstrate that 48 hr after infection the MP is strongly associated with ER.
Treatment of microsomes with proteinase K confirmed that erGFP is luminal, while MP was rapidly degraded in the presence and absence of detergent. When isolated membranes were washed with agents that are known to remove peripheral membrane proteins, the MP remained associated with the microsome vesicles and was dissociated only in the presence of nonionic detergent (Fig. 5). Taking these results together, it appears that MP behaves as an integral ER membrane protein and that the MP affinity for ER membrane is largely hydrophobic in nature. Since the anti-MP antibody used in these studies reacts with a C-terminal sequence of the MP (32), we conclude that at least the C terminus resides on the cytosolic face of the microsome vesicles.
Additional studies are necessary to characterize the topology of the insertion of MP in the ER membrane in greater detail. In an earlier study it was suggested that two putative membrane-spanning regions in the MP are responsible for attachment to membranes (17). The association of MP with ER early in infection and its impact on the morphology of ER indicates an involvement of this cellular compartment in intracellular transport of MP as well as intercellular spread of virus infection. A role of ER in cell–cell communication has been suggested in an earlier study; Grabski et al. (44) reported intercellular partitioning of fluorescent probes through Pd by using probes that reside exclusively in the ER. It will be interesting to investigate the involvement of ER in intra- and intercellular movement in greater detail.
We propose a model for intracellular movement of MP as well as for cell–cell spread of TMV infection based on transport of the RNA or viral ribonucleoprotein complex containing MP via the ER. In this model MP in conjunction with the complex would be laterally distributed toward the Pd and translocated to neighboring cells on the basis of movement of the continuous ER membrane. As the association of MP with microtubules occurs during late stages of infection, it is possible that microtubules are involved in transport of MP-GFP away from the ER and thus may play a role in degradation of the MP as was suggested for the tobamovirus Ob (35). In earlier studies microtubule association of MP was discussed as playing a central role in cell–cell spread (18, 19). Further studies will help to determine the role of MP association with cellular components as the endomembrane system and the cytoskeleton in cell–cell spread of virus infection.
Acknowledgments
We thank Dr. Hal S. Padgett (The Scripps Research Institute) for helpful discussions and Drs. Csilla A. Fenczik (The Scripps Research Institute) and Sandy G. Lazarowitz (Cornell University, Ithaca, NY) for critical reading of the manuscript. We also thank Dr. David Baulcombe (John Innes Center, Norwich, U.K.) for the tobacco line with an ER-targeted GFP, Drs. Bimei Hong and Jeffrey Harper (The Scripps Research Institute) for the ATPase antibody, and Dr. Rebecca Boston (North Carolina State University, Raleigh) for the BiP antibody. C.R. was supported by Fellowship Re 1222/1-1 of the Deutsche Forschungsgemeinschaft. Other support was provided by National Science Foundation Grant MCB 9631124 and by The Scripps Family Chair.
ABBREVIATIONS
- Pd
plasmodesmata
- ER
endoplasmic reticulum
- TMV
tobacco mosaic virus
- MP
movement protein
- CP
coat protein
- GFP
green fluorescent protein
- erGFP
ER-targeted GFP
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- BiP
immunoglobulin light chain binding protein
References
- 1.Mezitt L A, Lucas W J. Plant Mol Biol. 1996;32:251–273. doi: 10.1007/BF00039385. [DOI] [PubMed] [Google Scholar]
- 2.McLean B G, Hempel F D, Zambryski P C. Plant Cell. 1997;9:1043–1054. doi: 10.1105/tpc.9.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staehelin L A. Plant J. 1997;11:1151–1165. doi: 10.1046/j.1365-313x.1997.11061151.x. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe Y, Emori Y, Ooshika I, Meshi T, Ohno T, Okada Y. Virology. 1984;133:18–24. doi: 10.1016/0042-6822(84)90421-5. [DOI] [PubMed] [Google Scholar]
- 5.Deom C M, Oliver M J, Beachy R N. Science. 1987;237:389–394. doi: 10.1126/science.237.4813.389. [DOI] [PubMed] [Google Scholar]
- 6.Meshi T, Watanabe Y, Saito T, Sugimoto A, Maeda T, Okada Y. EMBO J. 1987;6:2557–2563. doi: 10.1002/j.1460-2075.1987.tb02544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilf M E, Dawson W O. Virology. 1993;193:106–114. doi: 10.1006/viro.1993.1107. [DOI] [PubMed] [Google Scholar]
- 8.Tomenius K, Clapham D, Meshi T. Virology. 1987;160:363–371. doi: 10.1016/0042-6822(87)90007-9. [DOI] [PubMed] [Google Scholar]
- 9.Atkins D, Hull R, Wells B, Roberts K, Moore P, Beachy R N. J Gen Virol. 1991;72:209–211. doi: 10.1099/0022-1317-72-1-209. [DOI] [PubMed] [Google Scholar]
- 10.Oparka K J, Prior D A M, Santa Cruz S, Padgett H S, Beachy R N. Plant J. 1997;12:781–789. doi: 10.1046/j.1365-313x.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- 11.Waigmann E, Lucas W J, Citovsky V, Zambryski P. Proc Natl Acad Sci USA. 1994;91:1433–1437. doi: 10.1073/pnas.91.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf S, Deom C M, Beachy R N, Lucas W J. Science. 1989;246:377–379. doi: 10.1126/science.246.4928.377. [DOI] [PubMed] [Google Scholar]
- 13.Citovsky V, Knorr D, Schuster G, Zambryski P. Cell. 1990;60:637–647. doi: 10.1016/0092-8674(90)90667-4. [DOI] [PubMed] [Google Scholar]
- 14.Citovsky V, Wong M L, Shaw A L, Prasad B V V, Zambryski P. Plant Cell. 1992;4:397–411. doi: 10.1105/tpc.4.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper B, Dodds J A. J Gen Virol. 1995;76:3217–3221. doi: 10.1099/0022-1317-76-12-3217. [DOI] [PubMed] [Google Scholar]
- 16.Deom C M, Schubert K R, Wolf S, Holt C A, Lucas W J, Beachy R N. Proc Natl Acad Sci USA. 1990;87:3284–3288. doi: 10.1073/pnas.87.9.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore P J, Fenczik C A, Deom C M, Beachy R N. Protoplasma. 1992;170:115–127. [Google Scholar]
- 18.Heinlein M, Epel B L, Padgett H S, Beachy R N. Science. 1995;270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- 19.McLean B G, Zupan J, Zambryski P C. Plant Cell. 1995;7:2101–2114. doi: 10.1105/tpc.7.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beachy R N, Zaitlin M. Virology. 1975;63:84–97. doi: 10.1016/0042-6822(75)90373-6. [DOI] [PubMed] [Google Scholar]
- 21.Kolehmainen L, Zech H, von Wettstein D. J Cell Biol. 1965;25:77–97. doi: 10.1083/jcb.25.3.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osman T A M, Buck K W. J Virol. 1996;70:6227–6234. doi: 10.1128/jvi.70.9.6227-6234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shalla T A. J Cell Biol. 1964;21:253–264. doi: 10.1083/jcb.21.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Restrepo-Hartwig M A, Carrington J C. J Virol. 1994;68:2388–2397. doi: 10.1128/jvi.68.4.2388-2397.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Restrepo-Hartwig M A, Ahlquist P. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaad M C, Jensen P E, Carrington J C. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward B M, Medville R, Lazarowitz S G, Turgeon R. J Virol. 1997;71:3726–3733. doi: 10.1128/jvi.71.5.3726-3733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinlein M, Padgett H S, Gens J S, Pickard B G, Casper S J, Epel B L, Beachy R N. Plant Cell. 1998;10:1107–1120. doi: 10.1105/tpc.10.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt C A, Beachy R N. Virology. 1991;181:109–117. doi: 10.1016/0042-6822(91)90475-q. [DOI] [PubMed] [Google Scholar]
- 30.Kay R, Chan A, Daly M, McPherson J. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- 31.Haseloff J, Siemering K R, Prasher D C, Hodge S. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kahn T W, Lapidot M, Heinlein M, Reichel C, Cooper B, Gafny R, Beachy R N. Plant J. 1998;15:5–25. doi: 10.1046/j.1365-313x.1998.00172.x. [DOI] [PubMed] [Google Scholar]
- 33.Dewitt N D, Hong B, Sussman M R, Harper J F. Plant Physiol. 1996;112:833–844. doi: 10.1104/pp.112.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epel B L, Padgett H S, Heinlein M, Beachy R N. Gene. 1996;173:75–79. doi: 10.1016/0378-1119(95)00678-8. [DOI] [PubMed] [Google Scholar]
- 35.Padgett H S, Epel B L, Kahn T W, Heinlein M, Watanabe Y, Beachy R N. Plant J. 1996;10:1079–1088. doi: 10.1046/j.1365-313x.1996.10061079.x. [DOI] [PubMed] [Google Scholar]
- 36.Hepler P K. Eur J Cell Biol. 1981;26:102–110. [PubMed] [Google Scholar]
- 37.Knebel W, Quader H, Schnepf E. Eur J Cell Biol. 1990;52:328–340. [PubMed] [Google Scholar]
- 38.Quader H, Schnepf E. Protoplasma. 1986;131:250–252. [Google Scholar]
- 39.Heim R, Cubitt A B, Tsien R Y. Nature (London) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 40.Cubitt A B, Heim R, Adams S R, Boyd A E, Gross L A, Tsien R Y. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 41.Ormo M, Cubitt A B, Kallio K, Gross L A, Tsien R Y, Remington S J. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 42.Yang F, Moss L G, Phillips G N., Jr Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 43.Hepler P K, Palevitz B A, Lancelle S A, McCauley M M, Lichtscheidl I. J Cell Sci. 1990;96:355–373. [Google Scholar]
- 44.Grabski S, De Feijter A W, Schindler M. Plant Cell. 1993;5:25–38. doi: 10.1105/tpc.5.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]