Abstract
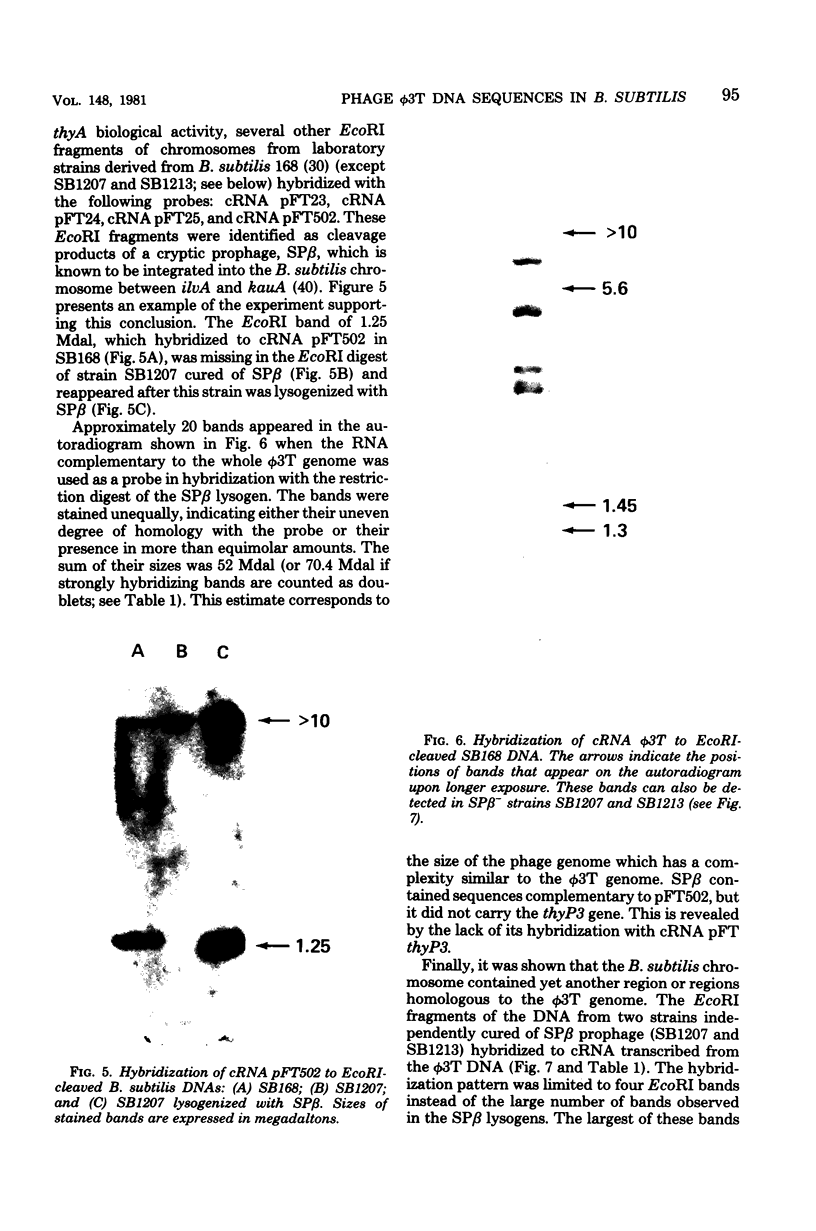
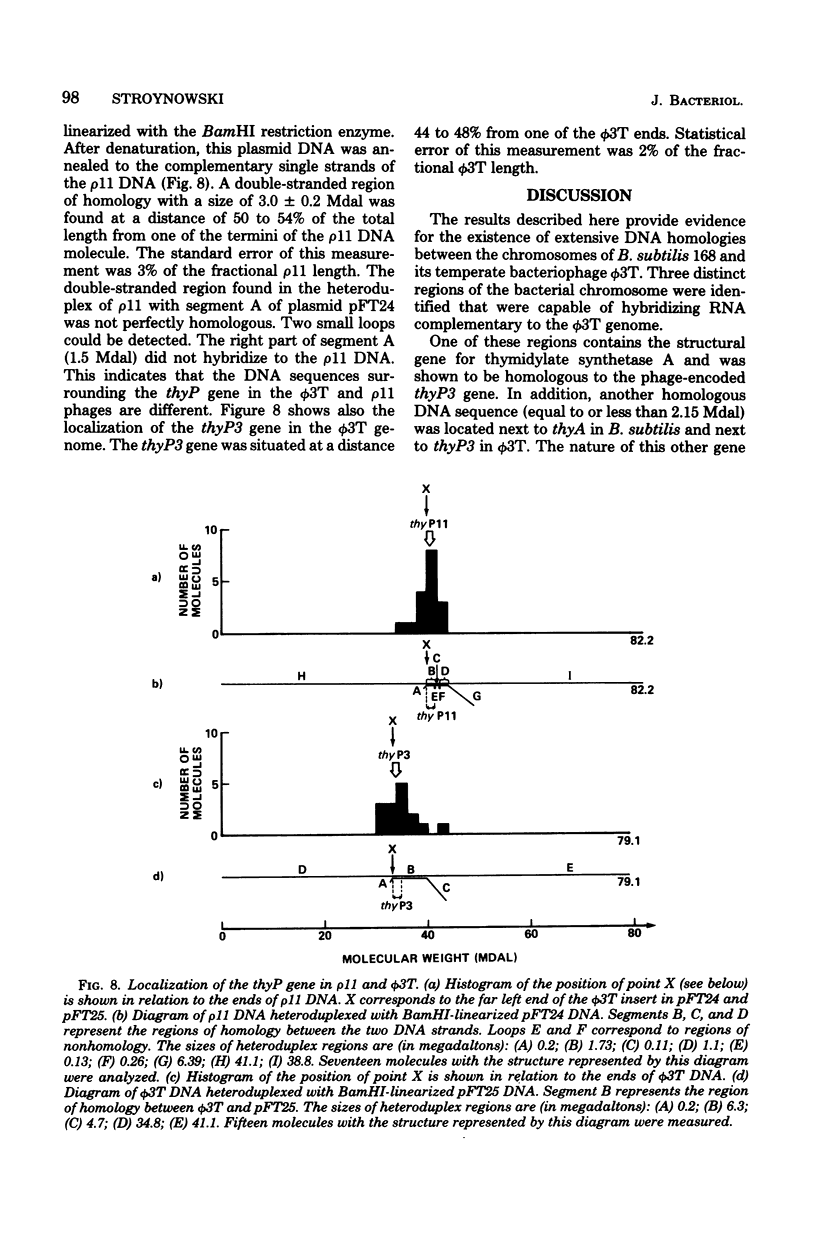
The Bacillus subtilis 168 chromosome was found to share extensive homology with the genome of bacteriophage phi 3T. At least three different regions of the bacterial genome hydridized to ribonucleic acid complementary to phi 3T deoxyribonucleic acid (DNA). The thymidylate synthetase gene, thyA, of B. subtilis and the sequences adjacent to it were shown to be homologous to the region in the phi 3T DNA containing the phage-encoded thymidylate synthetase gene, thyP3. SP beta, a temperate bacteriophage known to be integrated into the B. subtilis 168 chromosome, was demonstrated to be closely related to phi 3T. Other regions of the bacterial genome were also found to hybridize to the phi 3T probe. The nature and location of these sequences in the bacterial and phage chromosomes were not identified. It was shown however, that they were not homologous to either the thyP3 gene or the DNA surrounding the thyP3 gene. The chromosomes of other Bacillus species were also screened for the presence of phi 3T homologous sequences, and the thyP3 gene was localized in the linear genomes of phages phi 3T and rho 11 by heteroduplex mapping. It is suggested that the presence of sequences of phage origin in the B. subtilis 168 chromosome might contribute to the restructuring and evolution of the viral and bacterial DNAs.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arwert F., Rutberg L. Restriction and modification in Bacillus subtilis. Induction of a modifying activity in Bacillus subtilis 168. Mol Gen Genet. 1974;133(2):175–177. doi: 10.1007/BF00264838. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. H., Orrego J. C., Hutchison K. W., Halvorson H. O. New temperate bacteriophage for Bacillus subtilis, rho 11. J Virol. 1976 Nov;20(2):509–519. doi: 10.1128/jvi.20.2.509-519.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D., Bursztyn-Pettegrew H., Stroynowski I., Lederberg J. Expression of the thymidylate synthetase gene of the Bacillus subtilis bacteriophage Phi-3-T in Escherichia coli. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4145–4149. doi: 10.1073/pnas.73.11.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. M., Shoyab M., Baluda M. A. Studies on characterization of the integration sites of avian RNA tumor virus-specific DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):1005–1007. doi: 10.1101/sqb.1974.039.01.115. [DOI] [PubMed] [Google Scholar]
- Fujinaga K., Sekikawa K., Yamazaki H., Green M. Analysis of multiple viral genome fragments in adenovirus 7-transformed hamster cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):633–636. doi: 10.1101/sqb.1974.039.01.076. [DOI] [PubMed] [Google Scholar]
- Graham R. S., Young F. E., Wilson G. A. Effect of site-specific endonuclease digestion on the thyP3 gene of bacteriophage phi 3T and the thyP11 gene of bacteriophage rho11. Gene. 1977 Mar;1(2):169–180. doi: 10.1016/0378-1119(77)90027-0. [DOI] [PubMed] [Google Scholar]
- Günthert U., Pawlek B., Stutz J., Trautner T. A. Restriction and modification in Bacillus subtilis: inducibility of a DNA methylating activity in nonmodifying cells. J Virol. 1976 Oct;20(1):188–195. doi: 10.1128/jvi.20.1.188-195.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Nathans D. The genome of simian virus 40. Adv Virus Res. 1977;21:85–173. doi: 10.1016/s0065-3527(08)60762-9. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Davidson N. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology. 1974 Jan;57(1):93–111. doi: 10.1016/0042-6822(74)90111-1. [DOI] [PubMed] [Google Scholar]
- Klotz L. C., Zimm B. H. Size of DNA determined by viscoelastic measurements: results on bacteriophages, Bacillus subtilis and Escherichia coli. J Mol Biol. 1972 Dec 30;72(3):779–800. doi: 10.1016/0022-2836(72)90191-x. [DOI] [PubMed] [Google Scholar]
- Martin M. A., Khoury G., Fareed G. C. Specific reiteration of viral DNA sequences in mammalian cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):129–136. doi: 10.1101/sqb.1974.039.01.018. [DOI] [PubMed] [Google Scholar]
- Neiman P. E., Wright S. E., Purchase H. G. Studies of the interrelationship of chicken leukosis virus and host cell genomes by RNA-DNA hybridzation. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):875–883. doi: 10.1101/sqb.1974.039.01.102. [DOI] [PubMed] [Google Scholar]
- O'Donovan G. A., Neuhard J. Pyrimidine metabolism in microorganisms. Bacteriol Rev. 1970 Sep;34(3):278–343. doi: 10.1128/br.34.3.278-343.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Mudd J. A., Mangan J., Huang W. M., Subbaiah T. V., Marmur J. Properties of the defective phage of Bacillus subtilis. J Mol Biol. 1968 Jun 28;34(3):413–428. doi: 10.1016/0022-2836(68)90169-1. [DOI] [PubMed] [Google Scholar]
- Pellicer A., Wigler M., Axel R., Silverstein S. The transfer and stable integration of the HSV thymidine kinase gene into mouse cells. Cell. 1978 May;14(1):133–141. doi: 10.1016/0092-8674(78)90308-2. [DOI] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEAMAN E., TARMY E., MARMUR J. INDUCIBLE PHAGES OF BACILLUS SUBTILIS. Biochemistry. 1964 May;3:607–613. doi: 10.1021/bi00893a001. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Botchan M., Gallimore P., Ozanne B., Pettersson U., Williams J., Sharp P. A. Viral DNA sequences in cells transformed by simian virus 40, adenovirus type 2 and adenovirus type 5. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):615–632. doi: 10.1101/sqb.1974.039.01.075. [DOI] [PubMed] [Google Scholar]
- Sgaramella V., Ehrlich S. D., Bursztyn H., Lederberg J. Enhancement of transfecting activity of bacteriophage P22 DNA upon exonucleolytic erosion. J Mol Biol. 1976 Aug 25;105(4):587–602. doi: 10.1016/0022-2836(76)90237-0. [DOI] [PubMed] [Google Scholar]
- Sinsheimer R. L. Recombinant DNA. Annu Rev Biochem. 1977;46:415–438. doi: 10.1146/annurev.bi.46.070177.002215. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. R. Physical heterogeneity among Bacillus subtilis deoxyribonucleic acid molecules carrying particular genetic markers. J Bacteriol. 1969 Jun;98(3):1239–1247. doi: 10.1128/jb.98.3.1239-1247.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroynowski I. T. Integration of the bacteriophage phi 3T-coded thymidylate synthetase gene into the Bacillus subtilis chromosome. J Bacteriol. 1981 Oct;148(1):101–108. doi: 10.1128/jb.148.1.101-108.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker R. G. Acquisition of thymidylate synthetase activity by a thymine-requiring mutant of Bacillus subtilis following infection by the temperate phage phi 3. J Gen Virol. 1969 Jun;4(4):489–504. doi: 10.1099/0022-1317-4-4-489. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Revel H. R. The genome of bacteriophage T4. Bacteriol Rev. 1976 Dec;40(4):847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin R. E. DNA repair in Bacillus subtilis. I. The presence of an inducible system. Mol Gen Genet. 1977 Jun 8;153(2):211–218. [PubMed] [Google Scholar]
- Zahler S. A., Korman R. Z., Rosenthal R., Hemphill H. E. Bacillus subtilis bacteriophage SPbeta: localization of the prophage attachment site, and specialized transduction. J Bacteriol. 1977 Jan;129(1):556–558. doi: 10.1128/jb.129.1.556-558.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]