Abstract
Activation of the mitotic checkpoint pathway in response to mitotic spindle damage in eukaryotic cells delays the exit from mitosis in an attempt to prevent chromosome missegregation. One component of this pathway, hsMad2, has been shown in mammalian cells to physically associate with components of a ubiquitin ligase activity (termed the anaphase promoting complex or APC) when the checkpoint is activated, thereby preventing the degradation of inhibitors of the mitotic exit machinery. In the present report, we demonstrate that the inhibitory association between Mad2 and the APC component Cdc27 also takes place transiently during the early stages of a normal mitosis and is lost before mitotic exit. We also show that Mad2 associates with the APC regulatory protein p55Cdc in mammalian cells as has been reported in yeast. In contrast, however, this complex is present only in nocodazole-arrested or early mitotic cells and is associated with the APC as a Mad2/p55Cdc/Cdc27 ternary complex. Evidence for a Mad2/Cdc27 complex that forms independent of p55Cdc also is presented. These results suggest a model for the regulation of the APC by Mad2 and may explain how the spindle assembly checkpoint apparatus controls the timing of mitosis under normal growth conditions.
The accurate segregation of sister chromatids to two daughter cells during mitosis depends on the proper attachment of the kinetochores to a bipolar mitotic spindle apparatus. Failures in this attachment process result in unattached kinetochores at metaphase and chromosomes that are not under tension; both events are sensed by the cell and can send signals to prevent the exit from mitosis (for reviews see ref. 1). The sensors and signal transduction pathway involved in this response are referred to as the spindle assembly (or mitotic) checkpoint mechanism.
A great deal of progress has been made recently in understanding the molecular mechanisms involved in the activation of the spindle assembly checkpoint in eukaryotic cells. Based on the pioneering studies in budding yeast, at least seven genes are required for the execution of the spindle assembly checkpoint in response to mitotic spindle inhibitors, namely, MAD1–3 (3), BUB1–3 (2), and MPS1 (3). It is known that Mad1 in yeast becomes hyperphosphorylated in response to spindle damage and that this phosphorylation depends on the presence of Bub1, Bub3, and Mad2 (4). Overexpression of Mps1 causes constitutive phosphorylation of Mad1 and a delay in the onset of anaphase similar to that observed when the spindle is damaged (5). Interestingly, however, this delay is still dependent on Bub1, Bub3, and Mad2, demonstrating that these gene products are also required for events downstream of Mad1 phosphorylation. Bub1 (also a protein kinase) forms a complex with Bub3, and a dominant gain of function mutation in the Bub1 kinase domain has been shown recently to lead to a mitotic delay in the absence of spindle damage and Mad1 phosphorylation (6). The detailed nature of the complexes that form between the mitotic checkpoint genes and the mechanism of sensing and signaling spindle damage are still not well understood.
Vertebrate homologues of three of the mitotic checkpoint genes have been identified (MAD1, MAD2, BUB1) and all have been shown to be required for the execution of the mitotic checkpoint (7–10). Immunolocalization in mammalian cells has demonstrated that a subfraction of both Mad2 and Bub1 proteins are localized to the kinetochores of condensed chromosomes in prometaphase but are either displaced or masked upon spindle attachment. Diffuse Mad2 and Bub1 staining also is observed in the nucleoplasm (7, 9). In contrast, Mad1 is found at the kinetochore during interphase but localizes to the centrosome in prometaphase/metaphase (10). The localization and the timing of the formation of the proposed Mad1/Mad2 complex therefore are still unclear.
One obvious target for Mad2, considering its ability to inhibit the mitotic exit machinery, is the ubiquitin ligase referred to as the anaphase promoting complex (APC). The APC is a large multisubunit complex that ubiquitinates key cell cycle regulatory proteins that are targeted for degradation by the 26S proteosome to allow the exit from mitosis (11–13). Regulators of the APC have been identified that confer specificity to the ubiquitination reaction: Cdc20 in Saccharomyces cerevisiae (called Slp1 in Schizosaccharomyces pombe and p55Cdc in vertebrates) has been shown to target Pds1, an inhibitor of anaphase (14) and Cdh1/Hct1 is required for the degradation of cyclin B and Ase1 (15, 16). It has been shown recently that human Mad2 (hsMad2) can associate with the APC in HeLa cells when the mitotic spindle is disrupted by nocodazole treatment but that this association is lost upon release from nocodazole and before the degradation of cyclin B (17). The association between Mad2 and the APC also was shown to inhibit the ability of the APC to ubiquitinate cyclin B in a Xenopus extract. This model for the inhibitory role of Mad2 is consistent with the observations in S. pombe that overexpression of MAD2 causes a metaphase arrest and that weakened alleles of the APC components cut9 and nuc2 are hypersensitive to MAD2 overexpression (18).
It has been shown recently, both in fission and budding yeast, that Mad2 targets the APC regulator Slp1/Cdc20 (19, 20). Constitutive activation of the spindle assembly checkpoint by overexpression of Mad2 (in fission yeast) or Mps1 (in budding yeast) is relieved by mutations in Slp1/Cdc20 that block its association with Mad2. This suggests the possibility that in higher eukaryotes, the observed cell cycle-regulated Mad2/APC interaction may be mediated by the human homologue of Cdc20/Slp1 (called p55Cdc). Importantly, however, in budding yeast, it has been reported that the Mad2/Cdc20 interaction takes place throughout the cell cycle independent of the activation of the spindle assembly checkpoint. This prompted a more detailed analysis of the Mad2/p55Cdc/APC interaction in the cell cycle of higher eukaryotic cells reported here.
Finally, several lines of evidence suggest that the mitotic checkpoint genes are required to maintain the proper timing of mitosis and genomic stability in the absence of spindle inhibitors: mad1 and mad2 mutant strains of yeast lose chromosomes at higher rates than wild-type cells (21); cut4 and cut9 mutations in S. pombe, which diminish the activity of the APC, are suppressed by a deletion of mad2 (19); expression of a dominant-negative version of BUB1 in mammalian cells causes premature exit from mitosis under normal growth conditions (9); and chromosome instability in human tumor cell lines has been correlated with a defective response to nocodazole (22). In the present study, Mad2 is observed to associate with components of the APC and p55Cdc in the early phases of mitosis under normal growth conditions, providing a potential molecular explanation for the requirement of the mitotic checkpoint to maintain chromosome stability in cycling cells.
MATERIALS AND METHODS
Generation of Antibodies Against Human Cdc27 Protein.
The C-terminal six-tetratricopeptide repeat of Cdc27 (23) fused to a six-histidine tag was expressed in Escherichia coli from the expression vector pSTU68. Rabbit polyclonal antibodies against this form of Cdc27 were generated by injecting the fusion protein into two New Zealand rabbits as described (23). The sera of both rabbits were shown to specifically recognize Cdc27 in immunoprecipitation assays when compared with the sera of the same rabbits obtained before immunization.
Cell Culture and Synchronization.
HeLa cells were maintained in DMEM containing high glucose supplemented with antibiotics (100 units/ml penicillin-G/100 μg/ml streptomycin) and 10% fetal bovine serum. Cells were arrested at the G1/S boundary with 2 mM hydroxyurea for 12 hr and in metaphase with 100 ng/ml nocodazole for 16 hr. For the double-thymidine block and release experiments, cells were arrested for 14 hr with 2 mM thymidine, washed twice, released for 10 hr in medium without thymidine, arrested again for 14 hr with 2 mM thymidine, washed twice, and released into fresh medium. Samples were taken at the indicated time points.
Flow Cytometry.
Flow cytometric DNA quantification was done on cell nuclei. Samples were prepared according to refs. 24 and 25. Fluorescence was measured by using a Becton Dickinson FACScan (FACS Caliber) and analyzed by using multicycle (Phoenix Flow).
Immunoprecipitation and Immunoblotting.
HeLa cell extracts were prepared essentially as described (7). In short, cells were lysed in lysis buffer (50 mM Tris, pH 7.5/150 mM NaCl/1% Nonidet P-40/10% glycerol/2 mM EDTA) with protease and phosphatase inhibitors (50 mM NaF/0.1 mM orthovanadate/15 mM phenylmethylsulfonyl fluoride/15 mM 4-nitrophenyl phosphate/40 μg/ml aprotinin/20 μg/ml leupeptin). The extracts were cleared by centrifugation once for 5 min at 14,000 rpm. For Cdc27 and p55Cdc immunoprecipitations, 1 μl antiserum was used per 150 μg protein extract, and for Mad2 immunoprecipitations, 2 μl antiserum was used. Unless otherwise indicated, 60–100 μg protein extract was taken for direct immunoprecipitations, and 1–2 mg protein extract was taken for coimmunoprecipitations. The following antibodies were used: for Mad2 immunoprecipitation, the polyclonal rabbit antiserum against Mad2 (7); for Cdc27, the polyclonal rabbit antiserum described above; and for p55Cdc, the p55Cdc polyclonal antiserum from goat (Santa Cruz). The extracts were incubated for 2 hr with the antiserum before adding a 1:1 mix of Protein A and Gamma-bind G Sepharose beads (Pharmacia) for another hour. The beads were washed four times with wash buffer (50 mM Tris, pH 7.5/150 mM NaCl/1% Nonidet P-40/10% glycerol/2 mM EDTA/50 mM NaF) and three times with 50 mM Tris, pH 7.5, before boiling in sample buffer (1% SDS/50 mM Tris, pH 6.8/10% glycerol/0.01% Bromophenol blue).
p55Cdc and Mad2 protein was detected on 11.5% discontinuous SDS/polyacrylamide gels; Cdc27 protein was analyzed on 7.5% gels. Western blots were done essentially as described in Li and Benezra (7), except that poly(vinylidene difluoride) membrane (Immobilon-P, Millipore) was used and the signal was visualized by ECL plus (Amersham) as recommended by the manufacturer. For the Cdc27 Western analysis, the affinity-purified Cdc27 antibody described in Tugendreich et al. (23) was used at a 1:3,000 dilution. Mad2 antiserum and p55Cdc antiserum were diluted 1:500 and 1:1,000, respectively.
Immunodepletion Assays.
For Mad2 immunodepletion, immunoprecipitation was performed as described above, except that either 20 μl polyclonal Mad2 antiserum or Mad2 preimmune serum was added to 3.3 mg protein extract. For Cdc27, either 16 μl polyclonal Cdc27 antiserum or 16 μl Cdc27 preimmune serum was added to 1 mg protein extract. The extracts were incubated with Protein A and Gamma-bind G Sepharose beads. The depletion was repeated a second time with the same amount of antiserum and incubated overnight with Mad2 antiserum and 2 hr with Cdc27 antiserum, followed by incubation with Protein A and Gamma-bind G Sepharose beads (three times, 1 hr each). The beads were pooled and washed four times in wash buffer and three times with 50 mM Tris, pH 7.5.
For Mad2 immunodepletion one-tenth of the beads was boiled in sample buffer as described above. The final supernatants of the first immunoprecipitation were filtered through empty Micro Bio-Spin Columns (Bio-Rad). One-tenth of the final supernatants was used for immunoprecipitation with the Mad2 antibody to control for the efficiency of the immunodepletion (2 μl antiserum per 150 μg protein extract). The remaining supernatants (derived from 3 mg protein extract) were used for immunoprecipitation with Cdc27 antibody (1 μl antiserum per 150 μg protein extract).
For Cdc27 immunodepletion assays, the supernatants of the first immunoprecipitation were split into two aliquots and used for a second immunoprecipitation with either p55Cdc or Cdc27 antiserum as described above.
In Vitro Kinase Assays.
Protein extract (150 μg) was used for cyclin B immunoprecipitation (Santa Cruz, cyclin B1 antiserum) as described above. Immunoprecipitates were washed three times in wash buffer and three times in 25 mM Hepes, pH 7.4. The kinase reactions were done in 25 mM Hepes, pH 7.4/15 mM MgCl2/80 mM EGTA/1 mM DTT/0.1 mM ATP/3 μCi of [γ-32P]ATP. Histone H1 was used as a substrate. The reactions were incubated 20 min at 30°C, and the samples were boiled in sample buffer and analyzed by SDS/PAGE on a 15% discontinuous SDS/polyacrylamide gel. Band intensities were quantitated by PhosphorImager analysis.
RESULTS AND DISCUSSION
Mad2 Associates with Cdc27 in Cycling Cells During Mitosis.
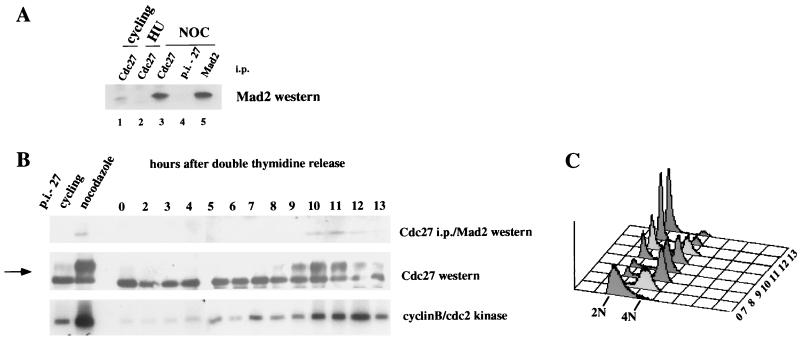
We first sought to determine whether Mad2 is in association with the APC in mammalian cells that were not exposed to mitotic spindle inhibitors. As shown in Fig. 1A, Mad2 is coimmunoprecipitated with the APC component Cdc27 in HeLa cells growing asynchronously in culture as well as in nocodazole-arrested cells (compare lanes 1, 3, and 4). No interaction is observed in cells arrested at the G1/S boundary with hydroxyurea (HU, lane 2) as reported previously (17). The association between Mad2 and Cdc27 is not quantitative as only a fraction of the total Mad2 pool is in this complex even in the nocodazole-arrested cells (see Fig. 1A legend and below).
Figure 1.
Mad2 transiently associates with Cdc27 during the cell cycle. (A) Mad2 is associated with Cdc27 in cycling cells. Extracts derived from cycling cells (lane 1), cells arrested with hydroxyurea (lane 2), or nocodazole (lane 3–5) were immunoprecipitated with the Cdc27 polyclonal antibody, and coimmunoprecipitating Mad2 protein was detected by Western blot analysis. Cdc27 preimmune serum was used as a negative control (lane 4), and the Mad2 polyclonal antiserum was used as a positive control (lane 5). Coimmunoprecipitations were done with 2 mg protein extract, and direct Mad2 immunoprecipitation was done with 100 μg protein extract (lane 5). (B) Association of Mad2 with Cdc27 is cell cycle-regulated. Cells were arrested in S phase by double-thymidine treatment and released. Protein extracts and nuclei (for FACS analysis) were prepared from samples taken at the indicated time points (in hours after the release). One milligram of protein extract was immunoprecipitated with Cdc27 antiserum followed by Western blot analysis of Mad2 (Top). Forty micrograms crude extract was used for the Cdc27 Western (Middle) and 150 μg for cyclin B/cdc2 kinase assays (Bottom). The arrow indicates the position of the phosphorylated forms of Cdc27. In all assays, extracts from cycling and nocodazole-arrested cells were included as indicated. For the coimmunoprecipitation assay, a Cdc27 preimmune control (p.i.-27) is shown. (C) FACS profile of the cell cycle release. The time points (in hours) are indicated.
To determine when during the cell cycle the Mad2/Cdc27 interaction takes place, HeLa cells were synchronized at the G1/S boundary with a double-thymidine block and the association of Cdc27 with Mad2 followed as a function of time after release. As shown in Fig. 1B, Cdc27 becomes hyperphosphorylated as the cells enter mitosis, and the association with Mad2 takes place at this time. A preferential association of Mad2 with hyperphosphorylated APC components has been observed previously (17) and was confirmed in this analysis (data not shown). Importantly, when the majority of the cells have exited mitosis (13 hr), the association of Cdc27 with Mad2 is diminished, after which time cyclin B/cdc2 kinase activity declines. These results demonstrate a physical interaction between Mad2 and components of the APC during a normal cell cycle.
p55Cdc Is Associated with Mad2 and Cdc27 in Nocodazole-Arrested Cells.
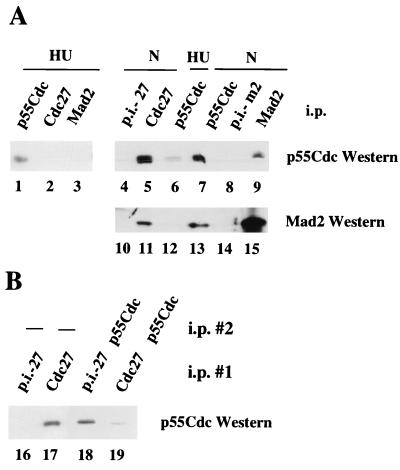
The association between Mad2 and the APC observed in immunoprecipitates does not need to be direct. Indeed, it has been shown in budding yeast that there is a physical interaction between Mad2 and the APC regulator Cdc20 (20), which may mediate the Mad2/APC interaction. However, the interaction between Mad2 and Cdc20 was found to vary only slightly throughout the cell cycle. To investigate this question in mammalian cells, extracts were prepared from cells arrested at the G1/S boundary with HU and in mitosis with nocodazole. As shown in Fig. 2A, antibodies to Mad2 coimmunoprecipitate p55Cdc (the Cdc20 homologue) in nocodazole-arrested cells but not in cells arrested in G1/S with hydroxyurea (compare lanes 3 and 9). As reported previously, we observe significantly less p55Cdc in G1 compared with mitotic cells (26), which may, in part, account for this difference (Fig. 2A, compare lanes 6 and 7). The association of Mad2 with p55Cdc in nocodazole-arrested cells but not in HU-arrested cells was confirmed first by immunoprecipitating with antibodies to p55Cdc followed by Western blotting with an anti-Mad2 antiserum (lanes 12 and 13). Note that approximately 30–40% of the p55Cdc found in nocodazole-arrested cells is found in a complex with Mad2 (Fig. 2A, compare lanes 7 and 9). Importantly, antibodies to Cdc27 coimmunoprecipitate the majority of the p55Cdc present in nocodazole-arrested cells (compare lanes 5 and 7) but none of the p55Cdc found in HU-arrested cells (compare lanes 1 and 2). Analysis of the supernatants after quantitative Cdc27 immunoprecipitation indicated about 80% of the total p55Cdc in nocodazole-arrested cells is complexed to Cdc27 (Fig. 2B, compare lanes 18 and 19).
Figure 2.
p55Cdc is associated with Mad2 and Cdc27 in nocodazole-arrested cells. Protein extracts (1.2 mg) derived from hydroxyurea-arrested cells (lanes 1–3 and 6) or nocodazole-arrested cells (lanes 4, 5, 7–9, 10, 11, and 13–15) were immunoprecipitated with the p55Cdc antibody (lanes 1, 6, 7, 12, and 13), with the Cdc27 antibody (lanes 2, 5, and 11), or the Mad2 antibody (lanes 3, 9, and 15). Preimmune controls were included (lanes 8 and 14, Mad2 preimmune serum, p.i.-m2; lanes 4 and 10, Cdc27 preimmune serum, p.i.-27). Western blots were probed with either the p55Cdc antibody (lanes 1–9) or the Mad2 antibody (lanes 10–15). (B) The majority of p55Cdc is found in association with Cdc27 in nocodazole-arrested cells. One milligram cell extract derived from nocodazole-arrested cells was immunodepleted with Cdc27 preimmune serum (lane 16) or with Cdc27 antiserum (lane 17). The supernatants of the first immunoprecipitation were split into two aliquots, and one aliquot was used for a second immunoprecipitation. Lanes 18, supernatant of lane 16, immunoprecipitated with p55Cdc antibody; lane 19, supernatant of lane 17, immunoprecipitated with p55Cdc antibody. All samples then were analyzed for p55Cdc by Western blotting. After immunodepletion with Cdc27 antiserum no Cdc27 protein was present in the supernatant (data not shown). Note that 1 mg protein extract was used for the first immunoprecipitation (lanes 16 and 17), and 500 μg protein extract was used for the second immunoprecipitation (lanes 18 and 19).
These results argue that the p55Cdc in association with Mad2 in nocodazole-arrested cells is likely to be in a ternary complex with Cdc27 (see also below). Some p55Cdc/Cdc27 complexes probably exist that are free of Mad2, but only a small amount of free p55Cdc/Mad2 complexes or free p55Cdc are likely to be present in nocodazole-arrested cells. Finally, since about only 10% of the Mad2 in nocodazole-arrested cells is in association with Cdc27 (Fig. 2, compare lanes 11 and 15; see also ref. 17), some free Mad2 may exist to perform other checkpoint functions.
p55Cdc, Mad2, and Cdc27 Form a Ternary Complex.
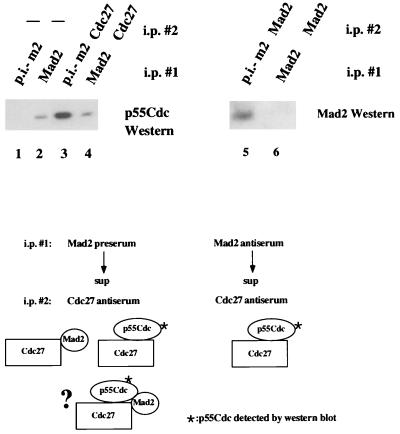
To confirm the existence of a ternary complex between Mad2, p55Cdc, and Cdc27, lysates from nocodazole-arrested cells first were immunodepleted with the anti-Mad2 antiserum or preimmune serum and then the supernatants were examined for Cdc27-p55Cdc complexes by coimmunoprecipitation/Western analysis. If a ternary complex between the three species exists, then the amount of p55Cdc immunoprecipitated with Cdc27 should be reduced when the first immunoprecipitation is performed with the Mad2 antiserum compared with the preimmune control (see model, Fig. 3). We first note that under the assay conditions employed, removal of Mad2 in the first immunoprecipitation is quantitative (Fig. 3, compare lanes 5 and 6). We then show that there is a significant reduction in the amount of p55Cdc associated with Cdc27 when the Mad2 antiserum is used in the first immunoprecipitation (compare lanes 3 and 4), confirming the existence of the ternary complex in vivo. Since immunoprecipitation with the anti-Mad2 antiserum does not quantitatively bring down the Cdc27/p55Cdc complexes, we conclude as above that some of the Cdc27/p55Cdc complexes are free of Mad2. This trivially could be due to the partial disruption of the complexes by the anti-Mad2 antibodies. Alternatively, it could be due to free APC/p55Cdc complexes being present in prometaphase cells or the presence of some Cdc27/p55Cdc that is not associated with the APC. We have not yet been able to quantitatively distinguish between these possibilities.
Figure 3.
Evidence for a trimeric Cdc27/Mad2/p55Cdc complex. Protein extracts derived from nocodazole-arrested cells were used in the following assays. Lanes: 1, one-step immunoprecipitation with preimmune Mad2 antiserum (p.i.-m2); 2, one-step immunoprecipitation with the Mad2 antiserum; 3, immunodepletion with Mad2 preimmune serum followed by Cdc27 immunoprecipitation of the supernatant; 4, immunodepletion with the Mad2 antiserum followed by Cdc27 immunoprecipitation of the supernatant. After immunodepletion with Mad2 antiserum, no Mad2 protein was present in the supernatant of the immunoprecipitation (lane 5 and 6). All samples were then analyzed for p55Cdc by Western blotting. The schematic shows the species predicted to contain p55Cdc after precipitating with the preimmune Mad2 antiserum (Left) or the anti-Mad2 antiserum in the first immunoprecipitation assuming both dimeric p55Cdc/Cdc27 and trimeric p55Cdc/Cdc27/Mad2 complexes exist. The decrease in the p55Cdc signal observed when the Mad2 antiserum is used relative to the preimmune control is a measure of the amount of ternary complex present in the extract. Note that 3 mg of protein was used for coimmunoprecipitations and 300 μg was used for direct immunoprecipitations.
Interactions of p55Cdc, Mad2, and Cdc27 During a Normal Mitosis.
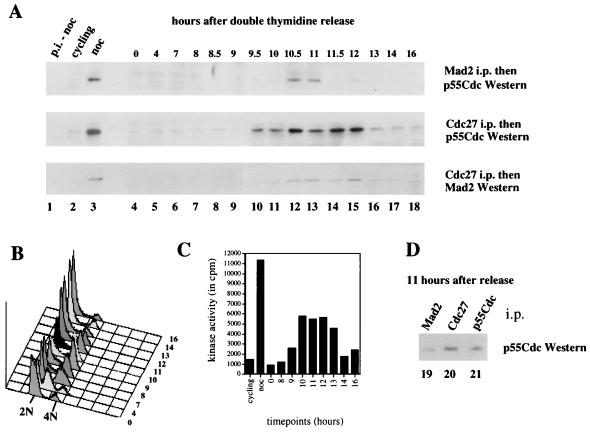
To follow the interactions between Mad2, p55Cdc, and Cdc27 during a normal cell cycle, HeLa cells were synchronized at the G1/S boundary by a double-thymidine block and then released as in Fig. 1B. The association of p55Cdc with Mad2 and Cdc27 was monitored by coimmunoprecipitation with either Mad2 or Cdc27 antibodies followed by Western blotting with antibodies to p55Cdc. As shown in Fig. 4, the majority of cells are in G2/M between 10 and 11 hr postrelease as assayed by fluorescence-activated cell sorter (FACS) analysis, and a peak of mitotic cyclin B kinase activity is observed at about 11 hr. At this time there is an association of p55Cdc with Mad2. This demonstrates that the Mad2/p55Cdc interaction takes place in a normal mitosis and not solely in response to nocodazole treatment (see above). The peak of this interaction takes place at about the peak of the observed association of p55Cdc with Cdc27 (Fig. 4A Middle). Importantly, as the cells proceed through mitosis (between 11 and 13 hr postrelease), the Mad2/p55Cdc interaction is lost approximately 1 hr before the dissociation of p55Cdc from Cdc27 (Fig. 4A, compare Top and Middle), suggesting the possibility that Mad2 leaves behind an active p55Cdc/Cdc27 complex. Note here that the majority of the immunoprecipitable p55Cdc in the cells 11 hr after the release is complexed to Cdc27 (compare lanes 20 and 21) as was observed in nocodazole-arrested cells. Finally, we observe that the Mad2/p55Cdc interaction is lost 1 hr before the dissociation of Mad2 from Cdc27 (Fig. 4A, compare Top and Bottom), strongly suggesting that at least some of the Mad2/Cdc27 complexes form independent of p55Cdc. After the Mad2/Cdc27 interaction is lost almost completely (14 hr postrelease, lane 17), cyclin B kinase activity drops significantly.
Figure 4.
Association of p55Cdc with Mad2 and Cdc27 during a normal cell cycle. Cells were arrested by a double-thymidine block and released as described in Materials and Methods. Samples were taken at the indicated time points (lanes 4–18), and protein extracts and nuclei (for FACS analysis) were prepared. (A) Coimmunoprecipitations. Extracts from cycling (lane 2) and nocodazole-arrested cells (lanes 1 and 3) were included as controls. Immunoprecipitations were done with either Mad2 antibody or Cdc27 antibody. The Mad2 immunoprecipitates were analyzed for coprecipitating p55Cdc (Top) and the Cdc27 immunoprecipitates were analyzed for coimmunoprecipitating p55Cdc (Middle) and Mad2 (Bottom) by Western analysis. The appropriate preimmune control assays were performed on extracts derived from nocodazole-arrested cells (lane 1, p.i.-noc). FACS analysis (B) and cdc2/cyclin B kinase assays (C) were performed at the indicated time points. (D) p55Cdc associated with Mad2 (lane 19) and Cdc27 (lane 20) as well as total p55Cdc in the cells (lane 21) 11 hr postrelease are shown. Five hundred micrograms protein extract was used for each immunoprecipitation.
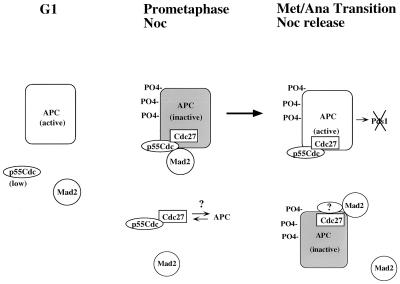
The results presented here suggest a model for the regulation of the APC by Mad2 diagrammed in Fig. 5. In G1 cells, there are low levels of p55Cdc and there is no association of this protein with Mad2 or components of the APC. The ability of the APC to degrade Pds1 in G1 (27) therefore is independent of an associated p55Cdc in this phase of the cell cycle perhaps because of differences in the phosphorylation status (28, 29) or some other structural state of the APC in G1 versus M that is reflected in dramatically different sedimentation behavior (17). As noted in Shiryama et al. (30) the stabilization of Pds1 in cdc20 mutants of S. cerevisiae is likely a result of changes in events of the previous mitosis as Cdc20 is undetectable in G1 in budding yeast. In the early stages of mitosis (or upon nocodazole arrest), Mad2 is associated with p55Cdc, which itself is nearly quantitatively complexed with Cdc27. We imagine that some or all of the Cdc27 is in a complex with other components of the APC. p55Cdc/Cdc27 complexes that do not contain Mad2 may exist (see above) but are either not associated with the APC, are inactivated by another mechanism, or cannot ubiquitinate key substrates because of its localization. Once the spindle assembly checkpoint is satisfied (either by progression through a normal cell cycle or after nocodazole release), Mad2 dissociates from p55Cdc, thereby allowing the APC/p55Cdc complex to participate in the ubiquitination of Pds1, whose degradation is required for the metaphase-to-anaphase transition (27). Interestingly, it appears that some Mad2 is in association with Cdc27 (and possibly the APC) after the Mad2/p55Cdc complex is no longer observed. This Mad2/APC complex may be required for the inhibition of ubiquitination of other non-p55Cdc-regulated substrates (such as cyclin B). This latter possibility must be tempered by the observation in yeast that the mitotic arrest observed in cells overexpressing Mad2 (or Mps1) is relieved by mutations that abrogate the Mad2/Cdc20(Slp1) interaction. If a Mad2/APC interaction is taking place in these cells independent of Cdc20(Slp1), then any inhibition of ubiquitination must not lead to cell cycle arrest. Interestingly, stabilization of cyclin B by mutations in cdh1 does not lead to complete loss of viability in S. cerevisiae (15, 16).
Figure 5.
Model for the regulation of the APC by Mad2. In G1 cells, the APC is capable of degrading Pds1 and cyclin B independent of an associated p55Cdc. No Mad2/p55Cdc or APC/p55Cdc complexes are detected in this phase of the cell cycle. In prometaphase and/or after checkpoint activation with the mitotic spindle inhibitor nocodazole (NOC), Mad2 and p55Cdc are complexed with the APC that is inactivated by virtue of the presence of Mad2. Free p55Cdc/Cdc27 complexes may exist in this phase of the cell cycle but, we speculate, are not part of the APC. At the metaphase-to-anaphase transition (or after nocodazole release), the Mad2/p55Cdc interaction is lost before the loss of the p55Cdc/APC interaction, allowing the latter complex to ubiquitinate the anaphase inhibitor Pds1. Mad2/Cdc27/APC complexes may exist at this time but are lost before the degradation of cyclin B (see text for details).
The tightly cell cycle-regulated interaction of Mad2 with p55Cdc/APC contrasts with observations in budding yeast in which the Mad2/cdc20 interaction was found to vary only slightly throughout the cell cycle (20). In addition, the reported failure to detect a Mad2/APC interaction in yeast implies that the Mad2/cdc20 complex fails to associate with the APC, which is in sharp contrast to the results reported here in mammalian cells. This could be because of inherent differences in the mechanism of Mad2 action between species. It is surprising, however, that a strong Mad2/cdc20 interaction is observed in G1 in budding yeast since Cdc20 protein has been reported to be undetectable in G1 in this organism (30).
Several key questions remain with regard to the Mad2/APC interaction data reported here and by others. First, it is important to determine whether it is the kinetochore-localized Mad2 that is in association with the APC/p55Cdc and whether this fraction of Mad2 is required for its checkpoint activity. The observation that a subfraction of two components of the APC (Tsg24, Cdc16) (31) and a subfraction of Mad2 are localized at the kinetochore in prometaphase is at least consistent with this possibility. Also, it is not yet known whether the release (or masking) of kinetochore-localized Mad2 upon spindle attachment is sufficient for the release of inhibition of the APC. This possibility seems somewhat unlikely if Mad2 is lost from the kinetochore upon spindle attachment given the coordinate nature of the metaphase-to-anaphase transition. Obviously, if the Mad2 epitope is simply masked by the incoming microtubule array, the dissociation from p55Cdc may be coordinated by some unknown mechanism before the metaphase-to-anaphase transition. Alternatively, the nucleoplasmic pool of Mad2 may be receiving signals from the checkpoint apparatus in response to incomplete spindle assembly, which leads to the association of Mad2 with p55Cdc and/or the subfraction of the APC, which has been observed on the mitotic spindle and centrosome (32). Prominent Mad2 staining on the centrosome or spindle has not been detected in HeLa cells (7, 8) but has sometimes been observed on these structures in PtK1 cells (33).
The observation that Mad2 can associate with components of the APC during a normal cell cycle may explain why defects in the checkpoint apparatus show cell cycle phenotypes in the absence of mitotic spindle inhibitors. In mammalian cells, the introduction of a dominant-negative version of BUB1 resulted in a more rapid exit from mitosis in synchronized cells relative to control cultures (9). In addition, loss of mitotic checkpoint control has been correlated with chromosome instability in human tumor cell lines (22). Finally, it has been shown recently that injection of Mad2 antibodies into mammalian cells in prometaphase causes a premature entry into anaphase (32, 33). From the data presented above, it is reasonable to propose that these observations result in part from the failure of Mad2 to interact with components of the APC during the early stages of mitosis and that this interaction is required to prevent the segregation of sister chromatids before the completion of the spindle assembly.
Acknowledgments
We are grateful to the members of the Benezra laboratory for helpful discussions, to G. Kirchweger and L. Michel for critical reading of the manuscript, and D. Domingo for FACS analysis. We thank J. Massague’s lab for antibodies and reagents and P. Hieter for the Cdc27 antibody and the Cdc27 expression plasmid. R.B. is supported by grants from the National Institutes of Health (GM54601) and the National Cancer Institute (CA08748). K.W. is the recipient of a Schroedinger Fellowship (J1561).
ABBREVIATIONS
- APC
anaphase promoting complex
- HU
hydroxyurea
- NOC
nocodazole
- FACS
fluorescence-activated cell sorter
Note
After submission of this manuscript, there have been two reports documenting the interactions between Mad2, p55Cdc, and the APC. Kallio et al. (1) and Fang et al. (34) demonstrate a Mad2/p55Cdc interaction that is cell cycle-regulated in agreement with our observations. Kallio et al. (1) also show that there is a detectable amount of Mad2 in association with the APC in the absence of p55Cdc in nocodazole-arrested cells, and we show that this Mad2/APC complex forms later in a normal mitosis. Fang et al. (34) reach a similar conclusion in cells released from the spindle assembly checkpoint. Finally, Fang et al. infer the existence of a ternary Mad2/p55Cdc/APC complex from binary immunoprecipitations, and a direct proof is provided in this report.
References
- 1.Kallio M, Weinstein J, Daum J R, Burke D J, Gorbsky G J. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoyt M A, Totis L, Roberts B T. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 3.Weiss E, Winey M. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardwick K G, Murray A W. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardwick K G, Weiss E, Luca F C, Winey M, Murray A W. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 6.Farr K A, Hoyt M A. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Benezra R. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 8.Chen R H, Waters J C, Salmon E D, Murray A W. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 9.Taylor S S, McKeon F. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 10.Jin D Y, Spencer F, Jeang K T. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 11.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King R W, Peters J M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 13.Zachariae W, Nasmyth K. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto A, Guacci V, Koshland D. J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwab M, Lutum A S, Seufert W. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 16.Visintin R, Prinz S, Amon A. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. Proc Natl Acad Sci USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X, Patterson T E, Sazer S. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S H, Lin D P, Matsumoto S, Kitazono A, Matsumoto T. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 20.Hwang L H, Lau L F, Smith D L, Mistrot C A, Hardwick K G, Hwang E S, Amon A, Murray A W. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 21.Li R, Murray A W. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 22.Cahill D P, Lengauer C, Yu J, Riggins G J, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Nature (London) 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 23.Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- 24.Nusse M, Beisker W, Hoffmann C, Tarnok A. Cytometry. 1990;11:813–821. doi: 10.1002/cyto.990110707. [DOI] [PubMed] [Google Scholar]
- 25.Giaretti W, Nusse M. Methods Cell Biol. 1994;41:389–400. doi: 10.1016/s0091-679x(08)61730-6. [DOI] [PubMed] [Google Scholar]
- 26.Weinstein J. J Biol Chem. 1997;272:28501–28511. doi: 10.1074/jbc.272.45.28501. [DOI] [PubMed] [Google Scholar]
- 27.Cohen-Fix O, Peters J M, Kirschner M W, Koshland D. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 28.Lahav-Baratz S, Sudakin V, Ruderman J V, Hershko A. Proc Natl Acad Sci USA. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters J M, King R W, Hoog C, Kirschner M W. Science. 1996;274:1199–1201. doi: 10.1126/science.274.5290.1199. [DOI] [PubMed] [Google Scholar]
- 30.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen P M, Brundell E, Starborg M, Hoog C. Mol Cell Biol. 1998;18:468–476. doi: 10.1128/mcb.18.1.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waters J C, Chen R H, Murray A W, Salmon E D. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorbsky G J, Chen R H, Murray A W. J Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang G, Yu H, Kirschner M W. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]