Abstract
Formation of small vesicles resembling synaptic vesicles can be reconstituted in vitro by incubating labeled homogenates of PC12 cells with ATP and two cytoplasmic proteins, AP3 and ARF1 [Faúndez, V., Horng, J.-T. & Kelly, R. B. (1998) Cell 93, 423–432]. To determine whether AP3 was mediating budding from plasma membranes or endosomes the organelle that generated the synaptic vesicles was characterized. The budding activity was enriched in organelles that labeled at 15°C, but not at 4°C, that excluded a marker of plasma membranes and that contained internalized transferrin, indicating that the precursor was an endosome. Vesicles formed from the endosomal precursor in vitro excluded transferrin. We conclude that ARF-mediated vesiculation into synaptic vesicle-sized organelles uses an endosomal precursor and occurs simultaneously in vitro with sorting of synaptic vesicle proteins from other membrane protein constituents of the endosome.
Membrane traffic between cellular organelles normally requires that cargo proteins be sequestered away from resident proteins into a membrane bud that vesiculates from the donor organelle to give a carrier vesicle. Both processes, vesiculation and membrane protein sorting, have been linked to a cytoplasmic coating mechanism that selects cargo while imparting curvature to the membrane (1). Sorting organelles such as the TGN or the endosome are hypothesized to use different coats for different cargoes.
To examine coating molecules involved in the sorting of synaptic proteins and their vesiculation from a precursor organelle into synaptic vesicle-sized structures (2), an in vitro reconstitution was developed using PC12 cells (3). The vesiculation process could be reconstituted in vitro with only two cytoplasmic proteins, the small GTPase ADP ribosylation factor (ARF1) (4) and the heterotetrameric adaptor protein complex AP3 (5). Left undefined was the nature of the precursor organelle that was vesiculating in vitro. In the reconstitution, the epitope-tagged, synaptic vesicle (SV) protein Vesicle-Associated Membrane Protein (VAMP) present in the precursor organelle was labeled by incubating transfected PC12 cells with radioactive antibody at 15°C (6, 7). At this temperature both plasma membrane and endosomes but not the SVs are labeled (3). Recently, it was shown that SVs in PC12 cells can arise from a plasma membrane compartment that is labeled at 15°C (8). We therefore examined whether the budding step reconstituted in our assay, which employs the adaptor complex, AP3, arises from a plasma membrane or an endosomal precursor.
Here we present evidence that the AP3 mediated vesiculation in vitro is from an endosomal organelle, not plasma membrane. Using membrane fractionation procedures, we were able to identify a transferrin (Tf) containing endosome, which is enriched in the SV protein VAMP and displays a high specific activity as a SV precursor. We also reexamined the link between vesiculation and sorting, motivated by recent data that vesiculation can be separated from sorting when pure coats are used to vesiculate protein-free liposomes (9). We find that sorting is indeed taking place in vitro during AP3-mediated vesiculation from PC12 endosomes, excluding Tf but not epitope-tagged VAMP from SVs. We conclude that AP3 and ARF1-mediated budding of SVs in PC12 cells is from an endosomal precursor.
MATERIALS AND METHODS
Materials.
125I and ECL reagents were obtained from Amersham. ATP, creatine phosphate, creatine kinase, BSA, and Sephadex G-50 were purchased from Boehringer Mannheim. Horseradish peroxidase (HRP)-conjugated rat Tf was obtained from Jackson ImmunoResearch and iron loaded following the method of Klausner et al. (10). Cell culture media and reagents were obtained from the University of California Cell Culture Facility (San Francisco), with the exception of Geneticin (G418), which was purchased from GIBCO/BRL. All other reagent grade materials were from Sigma or Fisher. Female Sprague–Dawley rats were from Bantin & Kingman (Fremont, CA).
Antibodies and Recombinant Proteins.
KT3 mAb against the T antigen (TAg) epitope tag was prepared as described in Grote et al. (6). HRP-conjugated goat anti-mouse and anti-rabbit IgG were from Cappel. Antibody against Na, K-ATPase (αF6), developed by Douglas M. Fambrough, was obtained from the Developmental Studies Hybridoma Bank (Iowa City, IA). Recombinant T31N mutant of ARF1 was purified from Escherichia coli expressing cDNA kindly provided by Dennis Shields (Albert Einstein College of Medicine, Bronx, NY) as described in Faúndez et al. (4).
Cell Culture.
All cell culture in this study was performed using the pheochromocytoma cell line, PC12, stably transfected with an epitope-tagged mutant of VAMP/synaptobrevin. In the mutant used (VAMP-TAg/N49A), asparagine in the conserved region of the cytoplasmic tail was replaced by alanine. This mutation was shown in previous studies to cause enhanced targeting to SVs (7) and thus is preferable for in vitro studies. Cells were grown and butyrated before the experiment as described in Faúndez et al. (4).
Cell Labeling.
VAMP-TAg/N49A PC12 cells were labeled with [125I]KT3 mAbs against the TAg epitope tag following the methods of Desnos et al. (3), described in detail in Clift-O’Grady et al. (11). Plasma membrane labeling by Tf-HRP involved the additional step of twice incubating the cells with 50 μM of the iron chelator deferoxamine mesylate in 150 mM NaCl, 2 mM CaCl2, 25 mM sodium acetate/acetic acid at pH 4.5 for 10 min on ice (12).
Subcellular Fractionation.
Labeled VAMP-TAg/N49A PC12 cells were pelleted and resuspended in budding buffer as described (11). Where indicated, a low-speed supernatant was generated from the homogenate by centrifugation at 1,000 × g for 5 min (5K g⋅m supernatant). In other cases, the 5K g⋅m supernatant was centrifuged for an additional 5 min at 10,000 × g to generate the fractions enriched in budding activity (50K g⋅m supernatant). In some cases, a high speed supernatant (950K g⋅m) was generated by centrifuging the 5K g⋅m or 50K g⋅m supernatants at 27,000 × g for 35 min. Between 200–250 μl (1–3 mg/ml) of the homogenate, 5K g⋅m, 50K g⋅m, or 950K g⋅m supernatants were layered on 10–45% sucrose gradients buffered with 20 mM Mops (pH 7.2), and prepared by using the Gradient Master (Biocomp Instruments, Fredericton, Canada). Sucrose gradients were centrifuged for 1 h at 116,000 × g at 4°C, or as indicated in a Beckman SW55 rotor. When experimental variables were to be compared, results were normalized to the same protein amounts for each of the samples. When homogenates were compared with the supernatants generated from them, the samples were normalized by volume. Sucrose refractive indices for each fraction were measured by a refractometer (Zeiss) and converted to percent sucrose. To identify SVs, 250 μl (1 mg/ml) of the 50K g⋅m or 950K g⋅m supernatants were layered on 5–25% glycerol velocity gradients and analyzed as described in Grote et al. (6). The cytosolic, unbound [125I]KT3 and Tf-HRP recovered on the top of the gradients was not included in the plots. All velocity centrifugations were performed in an SS34 rotor by using an RC2B centrifuge (DuPont-Sorvall).
In Vitro Budding Assay.
VAMP-TAg/N49A transfected PC12 cells were labeled as described above. In the standard reaction conditions, aliquots of 1 mg of homogenate, 5K g⋅m, or 50K g⋅m supernatants were incubated for 30 min at 37°C or 0°C, with an ATP-regenerating system (1 mM ATP/8 mM creatine phosphate/5 μg/ml creatine kinase) and 3 mg/ml rat brain cytosol prepared as described (3). When the effect of the T31N mutant was examined, the mixture was preincubated for 15 min at 0°C before warming. The tubes were chilled in ice for 5 min to stop the reaction before further treatments.
3′-3-Diaminobenzidine (DAB) Density Shift.
Density shift experiments were performed on cells labeled as described above, using DAB either in vivo or in vitro. In vivo: After stripping with deferoxamine mesylate, cells were incubated in the dark for 30 min at 0°C, with 1 mg/ml DAB (freshly prepared and filtered) in uptake buffer and 0.02% H2O2. Before scraping from culture dishes, the cells were washed once in uptake buffer. In vitro: 200–250 μl of 5K g⋅m or 50K g⋅m supernatants were incubated with 0.5 mg/ml DAB in budding buffer and 0.02% H2O2 at 0°C for 30 min in the dark with mild agitation. The reaction was stopped with the addition of NaN3 at a final concentration of 0.02%. In both cases, control reactions contained only H2O2.
Assays.
Tf-HRP: 75 μl of each gradient fraction were assayed for HRP activity in Falcon Microtest III 96-well tissue culture plates from Becton Dickinson by the addition of 115 μl of citrate buffer (50 mM citrate/100 mM PO4, pH 5.0) with O-phenylenediamine (tablets from Zymed) and 0.03% H2O2, prepared according to the manufacturer’s instructions. The assay was stopped after 7–25 min with the addition of 60 μl 2N H2SO4 to each well. Absorbance at 492 nm was detected using a Titertek Multiskan Plus by Flow Laboratories. HRP activity was found to be inhibited in a linear fashion by increasing concentrations of sucrose. At 45% sucrose, inhibition reached 60%, whereas no significant inhibition was observed at 10% sucrose. The data are not corrected for this inhibition, which de-emphasizes the recovery of Tf-HRP in fast sedimenting endosomes. Bio-Rad Protein Assay Dye Reagent was used for protein assays, BSA was used as standard.
RESULTS
Identification of the SV Precursor Organelle.
To study vesiculation and sorting of membrane proteins, we have used PC12 cells transfected with a VAMP/synaptobrevin construct encoding a VAMP with a TAg epitope tag on its C terminus. These transfectants allowed plasma membrane VAMP to be labeled with antibodies added to the extracellular medium (6). After incubating cells at 37°C, [125I]KT3 antibodies to the epitope tag were recovered in endosomes and in small vesicles similar in size, composition, and density to SVs (6, 7). Antibodies were not recovered in SVs when cells were labeled at 15°C but were found in a cytoplasmic organelle from which they could not be removed by acid stripping (3, 4). Incubation at 15°C also labeled an SV precursor that could generate labeled SVs when it was incubated in vitro at 37°C in the presence of ATP and cytosolic fractions (3). To determine if labeled plasma membrane or endosomal compartments were in vitro precursors of the SVs, we have made use of subcellular fractionation.
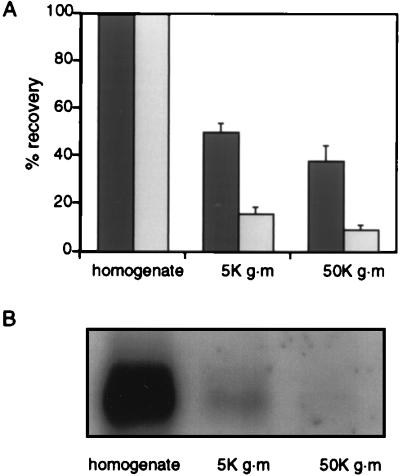
Compared with the entire homogenate, a significant percentage of the ability to generate labeled SVs was retained in a high-speed supernatant of the PC12 cell homogenate. Cells were labeled as described (3, 4) with [125I]KT3 antibody at 15°C, washed to remove unbound antibodies and then homogenized. Aliquots of the homogenate were centrifuged for 5 min at 1,000 × g to produce 5K g⋅m supernatants or subjected to an additional 10,000 × g to give 50K g⋅m supernatants and then compared for their ability to generate SVs during incubation at 37°C in the presence of ATP and brain cytosol. As in an earlier study (3), the postnuclear (5K g⋅m) supernatant retained significant budding activity (Fig. 1). On average 50.1 ± 3.5% of the labeled membranes in the 5K g⋅m supernatant could be recovered as SVs. The 50K g·m supernatant also retained budding activity (37.7 ± 6.5%) even though it contained only 9% of the labeled membranes that were present in the original homogenate, and <2% of the Na+K+ATPase, a plasma membrane marker, as measured by Western blotting (Fig. 1 A and B). Thus, almost half of the SV budding activity could be recovered in a labeled membrane fraction essentially devoid of labeled plasma membrane.
Figure 1.
Generation of SVs by subcellular fractions in vitro. (A) Comparison of efficiency of in vitro SV production from either homogenates, 5K g⋅m or 50K g⋅m supernatants. Aliquots of homogenates, 5K g⋅m (1,000 × g for 5 min) or 50K g⋅m (10,000 × g for 5 min) supernatants, were used to generate SVs in vitro. All reaction mixtures were then subjected to a 27,000 × g centrifugation for 35 min to remove the donor membranes. These high speed supernatants were analyzed on glycerol velocity gradients (5–25% glycerol in budding buffer; 218,000 × g for 75 min at 4°C), and the SVs generated in vitro were recovered in fractions 8–11. The dark shaded bars show the efficiency of budding activity from 5K g⋅m and 50K g⋅m supernatants as compared with the homogenate normalized to 100%. The light shaded bars show the percent recovery of labeled membranes, also compared with the homogenate normalized to 100%. An average of two different experiments is presented. To determine the amount of labeled membrane in each fraction, aliquots of [125I]KT3 labeled homogenates, 5K g⋅m or 50K g⋅m supernatants were subjected to a 27,000 × g centrifugation for 35 min to pellet membranes and to remove free cytosolic [125I]KT3. Resuspended membrane pellets were then counted for radioactivity. (B) Amounts of the plasma membrane protein Na, K-ATPase in PC12 cells homogenates were compared with those found in 5K g⋅m and 50K g⋅m supernatants. Membranes were pelleted by a 27,000 × g centrifugation for 35 min and their Na+,K+-ATPase content detected by using αF6 antibody after Western blot analysis.
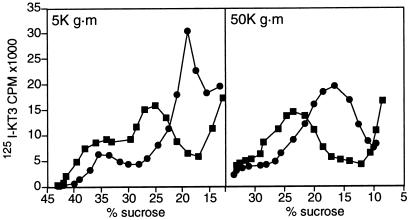
To characterize the SV precursor compartment more fully, 5K g⋅m and 50K g⋅m supernatants from [125I]KT3 labeled cells were analyzed by centrifugation on sucrose density gradients. The 5K g⋅m supernatant was resolved into two components (Fig. 2 Left, ▪) at 24 and 38% sucrose (summarized in Table 1). Centrifugation at 50K g⋅m depleted most of the fast sedimenting compartment from the supernatant and gave a single peak at 24% sucrose on sucrose density centrifugation (Fig. 2 Right, ▪; Table 1).
Figure 2.
Conversion of slow sedimenting endosomes to synaptic vesicles in vitro. (A) Endosomes were labeled by incubating PC12 cells expressing VAMP-TAg/N49A with [125I]KT3 for 40 min at 15°C and 5K g⋅m (Left) or 50K g⋅m (Right) supernatants were generated from homogenates. Incubation of the labeled supernatants at 37°C under standard reaction conditions (with ATP and rat brain cytosol for 30 min) caused a significant reduction of the slowly migrating endosomal peak, and a recovery of the label in a new compartment at approximately 17% sucrose, which is characteristic of SVs (•). Formation of SVs was inhibited when the supernatants were incubated at 0°C (■). The 5K g⋅m (Left) or 50K g⋅m supernatants (Right) were both subjected to a sucrose density centrifugation for 1 h at 116,000 × g, but the gradient in the right panel was shallower.
Table 1.
Separation of fast endosomes, slow endosomes and synaptic vesicles by nonequilibrium sucrose density gradient centrifugation
| Fraction | Slow endosomes
|
Fast endosomes
|
SVs
|
||
|---|---|---|---|---|---|
| [125I]KT3 | Tfn-HRP | [125I]KT3 | Tfn-HRP | [125I]KT3 | |
| 5K g⋅m supernatant∗ | 23.8 ± 0.3† (n = 14) | 24.7 ± 0.3 (n = 12) | 37.3 ± 0.6 (n = 12) | 37.6 ± 0.4 (n = 11) | |
| 50K g⋅m supernatant‡ | 22.4 ± 0.4 (n = 13) | 22.9 ± 0.8 (n = 6) | |||
| 950K g⋅m supernatant§ | 17.3 ± 0.3 (n = 12) | ||||
PC12 cells expressing VAMP-TAg/N49A were incubated for 40 min at 15°C with [125I] KT3 and Tf-HRP. Homogenates were then subjected to a 1,000 s × g centrifugation for 5 min (5K g⋅m).
Values of percent sucrose are an average of 1-4 peak points from each experiment. An average of several different experiments is shown ± SEM. All samples were centrifuged on 10-45% sucrose gradients for 1 hr at 116,0900 s × g.
5K g⋅m supernatants were subjected to an additional 10,000 × g centrifugation for 5 min and 50K g⋅m supernatants were analyzed.
5K g⋅m or 50K g⋅m supernatants were subjected to in vitro budding and analyzed on sucrose gradients. Following budding, [125I] KT3 but not Tf-HRP label was recovered in the SV peak.
The 38% and 24% components in a 5K g⋅m supernatant were not labeled detectably by incubating PC12 cells with [125I]KT3 antibodies at 4°C (data not shown). Because only the plasma membrane is believed to be labeled at 4°C, the lack of labeling is consistent with the paucity of plasma membrane in 5K g⋅m supernatants (Fig. 1B). The labeled compartments recovered at 38% and 24% thus have the characteristics of endosomes not plasma membrane. For convenience we call the two populations fast and slow endosomes to reflect the technique used to isolate them.
Because the 50K g⋅m supernatant contained much of the SV budding activity (Fig. 1A), we expected that the slow endosomes would be the precursor of SVs in vitro. This expectation was confirmed by incubating a 5K g⋅m supernatant under standard reaction conditions and analyzing the entire reaction mixture, which included both the product and the precursor membrane compartment, by sucrose gradient centrifugation (Fig. 2 Left •). Little of the 38% sucrose fast endosome peak was affected by the incubation at 37°C. The label found in the slow endosome peak was reduced, however, with the concomitant appearance of a peak at 17% sucrose, which is the position at which SVs are recovered (Table 1). Thus, the slow endosome fraction is the primary source of labeled SVs in this in vitro reconstitution assay, a conclusion confirmed by using the 50K g⋅m supernatant as a source of precursor membranes (Fig. 2 Right, •) and a shallower sucrose gradient. The budding from the slow endosomes is indistinguishable from that characterized earlier for PC12 homogenates (3). For example, the presence of the GDP-binding mutant of ARF1, ARF T31N, inhibited budding by 74.8% ± 2 (n = 2). We thus can conclude that labeled endosomal structures present in the 5K g.m and 50K g⋅m supernatant are precursors of SVs in vitro. Two endosomal fractions with similar densities could also be resolved by labeling PC12 cells with transferrin coupled to HRP at 15°C (Table 1). Labeling of these fractions by Tf-HRP did not occur at 0°C (data not shown). Analysis of the slow endosomes by whole-mount immunoelectron microscopy revealed tubular-vesicular organelles of 300–400 nm, similar in dimensions to peripheral endosomes labeled at 15°C in intact PC12 cells (H. de Wit, Y.L., R.B.K. & J. Klumperman, in preparation).
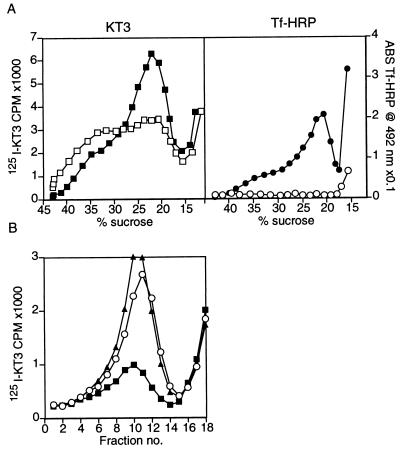
To determine if the Tf-HRP marker and the KT3 antibodies were indeed in the same endosomal fractions we employed the DAB density shift technique. Oxidation of DAB in compartments containing Tf-HRP crosslinks their contents, increases their density and reduces the HRP activity (13). By using two labeled protein markers, one of which is coupled to HRP, and DAB crosslinking, it is possible to localize markers to the same or to different intracellular compartments (see, for example: 14). In vitro DAB reactions performed on 50K g⋅m supernatants, resulted in a 53 ± 2% (n = 2) reduction in the [125I]KT3 label associated with the slow endosome fraction and in a shift in its density, suggesting that Tf-HRP reaches about half of these KT3-containing endosomes under the conditions used here (Fig. 3A Left). The DAB reaction caused the complete elimination of the Tf-HRP activity peak (Fig. 3A Right). Similar results were obtained when the DAB density shift reaction was performed on 5K g⋅m supernatants or if intact cells were treated in vivo with DAB before isolating the 5K g⋅m supernatants (data not shown). When in vitro budding reactions were carried out from 50K g⋅m supernatants that were first subjected to a DAB density shift, a 68 ± 0.4% (n = 2) decrease in budding efficiency was observed (Fig. 3B). This implies that the endosomes that are precursors of SVs in vitro contain both [125I]KT3 and Tf-HRP.
Figure 3.
Tf-HRP is present in the SV precursor endosomes. (A) Endosomes were generated by labeling PC12 cells expressing VAMP-TAg/N49A with both [125I]KT3 and Tf-HRP for 40 min at 15°C, followed by homogenization. 50K g⋅m supernatants were prepared and incubated with DAB and H2O2 for 30 min at 0°C. This caused an HRP catalyzed oxidation of DAB in the Tf-HRP containing organelles and a subsequent crosslinking of their contents. Control reactions were incubated under the same conditions with H2O2 only. Endosomes were then fractionated on sucrose gradients and analyzed for [125I]KT3 and Tf-HRP label. The DAB reaction caused a 53 ± 2% (n = 3) reduction in the KT3 label (Left) appearing at the slow endosomal peak (□), as compared with the control reaction (■). Tf-HRP activity (Right) was completely eliminated after the DAB reaction (○), compared with the control (•). (B) The same endosomes as in A were subjected to an in vitro budding reaction to produce SVs, after the DAB reaction. Membranes were then centrifuged for 35 min at 27,000 × g, and supernatants were analyzed on glycerol velocity gradients. The DAB reaction caused a 67% decrease in budding efficiency (■) as compared with the control (▴) that lacks DAB and H2O2 treatment. This demonstrates that [125I]KT3 is sorted to SVs from a Tf-HRP-containing compartment. When membranes were treated with H2O2 alone (○), no significant decrease in budding efficiency was detected. DAB alone also had no effect on the budding efficiency (data not shown).
Sorting from Endosomes in Vitro.
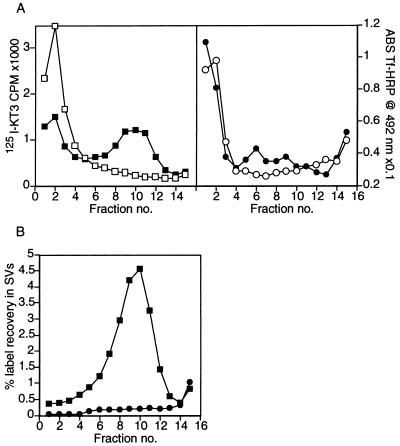
Previous studies of SV formation in vivo and in vitro had suggested that both vesiculation and membrane protein sorting were occurring (3, 15, 16, 17) but did not establish whether the processes were simultaneous. This issue needed to be considered because the slow endosomal precursor was found to be 2.7 ± 0.3 (n = 10) fold enriched in the ratio of KT3 antibody label to Tf-HRP activity, when compared with the non-precursor fast endosomal peak (the corrected value is 3.6-fold, when inhibition of HRP by sucrose is considered—see Materials and Methods). Such data could imply that sorting of SV membrane proteins occurs exclusively during the formation of the endosome precursor before the vesiculation of SVs. To determine whether this level of sorting was sufficient or whether sorting also occurred during the vesiculation step, endosomes labeled at 15°C with both [125I]KT3 and Tf-HRP were used to quantify sorting in vitro. To do this, the entire reaction mixture after in vitro incubation of slow endosomes was analyzed by a 5–25% glycerol velocity gradient sedimentation under standard conditions (3). The donor membranes before incubation were recovered in fractions 1 and 2, which correspond to the sucrose pad below the glycerol gradient (Fig. 4A Left, □). After incubation at 37°C, less of the [125I]KT3 label was recovered in the precursor on the sucrose pad, and a peak of radioactivity appeared in the position of SVs (fractions 9–11, Fig. 4A Left, ▪). In contrast, little of the Tf-HRP was recovered in the SV peak, even after incubation (Fig. 4A Right, •). Exclusion of transferrin from the newly-synthesized SV peak was confirmed by using [125I]Tf instead of Tf-HRP (data not shown). The exclusion of Tf-HRP from SVs could also be demonstrated by removing the precursor endosomes by differential centrifugation before glycerol gradient sedimentation (Fig. 4B). Recovery of the endosomal starting material in the SV peak was much higher for [125I]KT3 (▪) than for Tf-HRP (•). After in vitro formation of SVs from the slow endosome precursor, the recovery of [125I]KT3 in SVs was 16 ± 3 (n = 2) times higher than the recovery of Tf-HRP. SVs rich in KT3 antibodies but excluding Tf-HRP could also be observed when endosomal precursors were incubated with AP3 and ARF1 instead of brain cytosol (data not shown). Thus, SV formation from endosomes is associated with sorting of SV membrane proteins, in addition to a vesiculation step.
Figure 4.
[125I]KT3 is enriched in slowly sedimenting endosomes and sorted to newly formed SVs. (A) Labeled endosomes in a 50K g⋅m supernatant were incubated under standard reaction conditions at 37°C to generate labeled SVs. The entire reaction mixture was analyzed by sedimentation on a 5 to 25% glycerol velocity gradient, layered over a dense sucrose pad as described in Fig. 1A. A peak of [125I]KT3 label was recovered in fraction 10 (Left) corresponding to newly formed SVs (■). When this in vitro reaction was carried out at 0°C, no SVs were made and the endosome precursors accumulated on the sucrose pad at the foot of the glycerol gradient (□). Tf-HRP label (right) accumulated with the heavy membranes at the bottom of the gradient, whether the in vitro reaction was carried out at 37°C (•), or 0°C (○). This implies that Tf-HRP was not sorted into SVs. (B) Endosomes in a 50K g⋅m supernatant were isolated on sucrose gradients as in Fig. 2. An in vitro reaction was also carried out under standard reaction conditions on another fraction of this 50K g⋅m supernatant. When the reactions were completed, the reaction mixture was subjected to a 35 min centrifugation at 27,000 × g to pellet the donor membranes. After fractionation of the supernatant on glycerol velocity gradients, SVs were identified by a peak of [125I]KT3 in fractions 8–11 (■), characteristic of SVs. No Tf-HRP label was recovered at the SV peak position (•). Plots represent the recovery of label in SVs as a percent of the total label in the endosome donor.
DISCUSSION
The heterotetrameric proteins AP1 and AP2 have been linked to budding events from Golgi and from plasma membrane, respectively. In a previous study we had shown that AP3-mediated a budding step in vitro but had not identified the precursor. We now report that the budding step mediated by ARF1 and AP3 is from an endosome. The donor compartment has labeling properties, sedimentation characteristics, and morphology different from plasma membrane and is co-labeled by an endosomal marker. These data are consistent with the presence of AP3 on endosomes both by light (18) and by electron microscopy (19).
Genetic studies using yeast cells mutant in AP3 have provided evidence for budding from Golgi (23). Until this work, the only organelle that was known to generate SVs in PC12 cells was the plasma membrane. Schmidt et al. showed that SVs could be generated from plasma membrane associated structures in PC12 cells that accumulated at 18°C and excluded Tf receptors (8). The assay used in this study does not examine plasma membrane-derived budding because it uses a precursor depleted of plasma membrane by a low speed centrifugation step. Using a different in vitro assay, in which only the plasma membrane is labeled, SV-sized structures were generated by a coating mechanism that required neither AP3 nor ARF1 (G. Shi, V.F., J. Roos, E. C. Dell’Angelica, and R.B.K., unpublished work). The physiological significance of the alternative pathways of SV formation remains to be established.
It is likely that AP3 mediates only one of the pathways that originate from endosomes. Clathrin-coated vesicles have been seen in electron micrographs of endosomes (20). COPI, or a close homolog, is involved in transport from early to late endosomes (21, 22). AP3 has been reported to mediate a Golgi to vacuolar pathway in yeast (23, 24) and to be involved in pigment granule biogenesis (18, 25, 26). Both COPI and AP3 thus seem to have the capacity to mediate budding from both Golgi and endosomal membranes, although the details of the mechanisms involved may turn out not to be identical.
The nature of the two endosomal peaks resolved by density in 5K g·m supernatants is not yet completely defined. From electron micrography (de Wit, et al., unpublished work), the relatively slow sedimenting characteristics of the 24% sucrose endosomes cannot be attributed to unusually small size. Indeed they have the dimensions expected of endosomes. What then is the explanation of the “fast endosomes”? Perhaps the faster sedimenting endosomes, enriched in Tf-labeling at 15°C, are those that are migrating or have migrated to the perinuclear region. Their more rapid sedimentation characteristics could be attributed to fusion that forms interconnected endosomal structures or to association with the cytoskeleton.
Vesiculation was believed to be mediated by a recognition event between cargo proteins and adaptor complexes. Recently, however, we have learned that yeast COPII proteins can vesiculate cargo-free liposomes (9). It thus becomes imperative to distinguish vesiculation events mediated by cargo proteins from lipid-mediated ones. We have already shown that AP3 cannot coat SVs shorn of their exposed proteins by protease activity (5). Here we show that membrane protein sorting is occurring during the vesiculation process, a result confirmed by quantitative electron microscopy (DeWitt, et al., unpublished work). Thus the AP3 plus ARF1 coating mechanism is causing sorting as well as vesiculation in this particular in vitro reconstitution.
Genetic studies of Drosophila and mouse have implicated AP3 in the biogenesis of a diverse array of cytoplasmic storage granules, including pigment granules, platelet alpha-granules, and secretory granules in cytotoxic T cells (26). The data here show that AP3 is involved in retrieving selected membrane components from an endosome into a small vesicle. A way of combining a role for AP3 in storage organelle formation with one in endosomal membrane retrieval is suggested by experiments on endocrine secretory granule formation. Maturation of an immature endocrine secretory granule into a mature one involves the retrieval of a class of membrane proteins into small vesicles by an AP1-mediated process (27–30). Perhaps the aberrations in storage granule biogenesis in AP3 deficient mice and Drosophila are due to defects in granule maturation associated with inefficient removal of excess membrane from an endosome-related intermediate by AP3-mediated vesiculation.
Acknowledgments
The authors wish to thank Dr. Jim-Tong Horng for providing the VAMP-TAg/N49A PC12 cell line. The αF6 antibody developed by Douglas M. Fambrough was obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242, under contract NO1-HD-7-3263 from the National Institute of Child Health and Human Development. We thank Dennis Shields (Albert Einstein College of Medicine, Bronx, NY) for the ARF1 plasmids. We are also grateful for the technical assistance of Michael O’Grady and to Leslie Spector for her help in the preparation of the manuscript. This work was supported by grants to Regis B. Kelly from National Institutes of Health (NS09878 and DA10154) and a postdoctoral fellowship to C.D. from the Ministere Français de l’Enseignement Superieur et de la Recherche, France. V.F. is funded by a Fogarty Fellowship from the National Institutes of Health; Y.L. is funded by a Human Frontier Science Program Postdoctoral Fellowship.
ABBREVIATIONS
- ARF1
ADP ribosylation factor
- DAB
3′3-diaminobenzidine
- HRP
horseradish peroxidase
- SV
synaptic vesicle
- TAg
T-antigen
- Tf
transferrin
- VAMP
vesicle-associated membrane protein
- AP
adaptor protein
References
- 1.Kirchhausen T, Bonifacino J S, Reizman H. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 2.Clift-O’Grady L, Linstedt A D, Lowe A W, Grote E, Kelly R B. J Cell Biol. 1990;110:1693–1703. doi: 10.1083/jcb.110.5.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desnos C, Clift-O’Grady L, Kelly R B. J Cell Biol. 1995;130:1041–1049. doi: 10.1083/jcb.130.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faúndez V, Horng J-T, Kelly R B. J Cell Biol. 1997;138:505–515. doi: 10.1083/jcb.138.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faúndez V, Horng J-T, Kelly R B. Cell. 1998;93:423–432. doi: 10.1016/s0092-8674(00)81170-8. [DOI] [PubMed] [Google Scholar]
- 6.Grote E, Hao J C, Bennett M K, Kelly R B. Cell. 1995;81:581–589. doi: 10.1016/0092-8674(95)90079-9. [DOI] [PubMed] [Google Scholar]
- 7.Grote E, Kelly R B. J Cell Biol. 1996;132:537–549. doi: 10.1083/jcb.132.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt A, Hannah M J, Huttner W B. J Cell Biol. 1997;137:445–458. doi: 10.1083/jcb.137.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuoka K, Orci L, Amherdt M, Benarek S Y, Hamamoto S, Schekman P. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 10.Klausner R D, Van Renswoude J, Ashwell G, Kempf C, Schechter A N, Dean A, Bridges K R. J Biol Chem. 1983;258:4715–4724. [PubMed] [Google Scholar]
- 11.Clift-O’Grady, L., Desnos, C., Lichtenstein, Y., Faúndez, V., Horng, J.-T. & Kelly, R. B. (1998) Methods: A Companion to Methods in Enzymology 16, in press. [DOI] [PubMed]
- 12.Stoorvogel W, Strous G J, Geuze H J, Oorschot V, Schwartz A L. Cell. 1991;65:417–427. doi: 10.1016/0092-8674(91)90459-c. [DOI] [PubMed] [Google Scholar]
- 13.Courtoy P J, Quintart J, Baudhuin P. J Cell Biol. 1984;98:870–876. doi: 10.1083/jcb.98.3.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Weert A W M, Dunn K W, Geuze H J, Maxfield F R, Stoorvogel W. J Cell Biol. 1995;130:821–834. doi: 10.1083/jcb.130.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linstedt A D, Kelly R B. Neuron. 1991;7:309–317. doi: 10.1016/0896-6273(91)90269-6. [DOI] [PubMed] [Google Scholar]
- 16.Cameron P L, Südhof T C, Jahn R, De Camilli P. J Cell Biol. 1991;115:151–164. doi: 10.1083/jcb.115.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cameron, P., Mundigl, O. & De Camilli, P. (1993) J. Cell Sci. Suppl. 17, 93–100. [DOI] [PubMed]
- 18.Dell’Angelica E C, Ohno H, Ooi C E, Rabinovich E, Roche K W, Bonifacino J S. EMBO J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dell’Angelica E C, Klumperman J, Stoorvogel W, Bonifacino J S. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- 20.Stoorvogel W, Oorschot V, Geuze H J. J Cell Biol. 1996;132:21–33. doi: 10.1083/jcb.132.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruenberg J, Maxfield F R. Curr Opin Cell Biol. 1995;7:552–563. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 22.Whitney J A, Gomez M, Sheff D, Kreis T E, Mellman I. Cell. 1995;83:703–713. doi: 10.1016/0092-8674(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 23.Cowles C R, Odorizzi G, Payne G S, Emr S D. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- 24.Stepp J D, Huang K, Lemmon S K. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson F, Peden A A, Christopoulou L, Robinson M S. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odorizzi G, Cowles L R, Emr S D. Trends Cell Biol. 1998;8:282–288. doi: 10.1016/s0962-8924(98)01295-1. [DOI] [PubMed] [Google Scholar]
- 27.Grimes M, Kelly R B. J Cell Biol. 1992;117:531–538. doi: 10.1083/jcb.117.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dittie A S, Hajibagheri N, Tooze S A. J Cell Biol. 1996;132:523–536. doi: 10.1083/jcb.132.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milgrim S L, Kho S T, Martin F V, Mains R E, Eipper B A. J Cell Sci. 1997;110:695–706. doi: 10.1242/jcs.110.6.695. [DOI] [PubMed] [Google Scholar]
- 30.Klumperman J, Kuliawat R, Griffith J M, Geuze H J, Arvan P. J Cell Biol. 1998;141:359–371. doi: 10.1083/jcb.141.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]