Abstract
We sublocalized the yeast nucleoporin Nup82 to the cytoplasmic side of the nuclear pore complex (NPC) by immunoelectron microscopy. Moreover, by in vitro binding assays we showed that Nup82 interacts with the C-terminal region of Nup159, a yeast nucleoporin that previously was also localized to the cytoplasmic side of the NPC. Hence, the two nucleoporins, Nup82 and Nup159, form a cytoplasmically oriented subcomplex that is likely to be part of the fibers emanating from the cytoplasmic ring of the NPC. Overexpression of Rss1/Gle1, a putative nucleoporin and/or mRNA transport factor, was shown previously to partially rescue depletion of Nup159. We show here that overexpression of Rss1/Gle1 also partially rescued depletion of Nup82. Depletion of either Nup82, Nup159, or Rss1/Gle1 was shown previously to inhibit mRNA export. As was reported previously for depletion of Nup159 or of Rss1/Gle1, we show here that depletion of Nup82 has no detectable effect on classical nuclear localization sequence-mediated nuclear import. In summary, the nucleoporins Nup159 and Nup82 form a cytoplasmically oriented subcomplex of the NPC that is likely associated with Rss1/Gle1; this complex is essential for RNA export, but not for classical nuclear localization sequence-mediated nuclear protein import.
Keywords: nucleoporin 82/nucleoporin 159/binding assays/immunoelectron microscopy
Nuclear pore complexes (NPCs) are large, proteinaceous macromolecular complexes [≈120 MDa in vertebrates (1); ≈60 MDa in yeast (2)] spanning the nuclear envelope (NE), a double membrane whose lumen is continuous with the lumen of the endoplasmic reticulum (3). As the nuclear gate, the NPC is vital for regulating the transport of ribonucleoproteins (RNP) and proteins to and from the nucleus and plays a critical role in the formation and maintenance of nuclear structure.
Ultrastructural studies show that the NPC is an 8-fold radially symmetric structure, bilaterally symmetric with respect to the cytoplasm and the nucleus, which contains peripheral components unique to each face (4). Studies of amphibian NPCs show that the cytoplasmic face contains eight particles evenly spaced around a ring (4, 5) and eight fibrils 30–50 nm long (6, 7) extending into the cytoplasm. On the nucleoplasmic face of the NPC, so-called nuclear baskets or fishtraps are seen (7–9) that consist of eight fibrils that extend 100 nm from a ring at the nucleoplasmic face of the NPC and are connected by a narrower ring at their termini.
Of the more than 20 NPC proteins (called nucleoporins or Nups for short) identified so far in yeast, only a few have been sublocalized within the NPC. The nucleoporins Nsp1 (10) and Nic96 (Michael P. Rout, personal communication), which are found in a complex with Nup49 (11, 12), and Nup57 (13) have been localized to the cytoplasmic and nuclear sides of the NPC, so presumably this entire complex is found on both sides of the NPC. A second complex found on both sides of the NPC and connected to the above one contains Nup188, Nic96, and the pore membrane protein, Pom152 (14). Another nucleoporin, Nup170, which interacts genetically with Pom152, and a homolog of Nup170, Nup157, also are found on both sides of the NPC (15).
Nup159 is an essential nucleoporin (16, 17) that has been localized to the cytoplasmic face of the NPC (17). It has been shown to interact with nuclear transport factors and is required for nuclear export of poly(A)+ RNA but not for classical nuclear localization sequence (NLS)-mediated protein import. This study shows that Nup82, a protein required for poly(A)+ RNA export (18, 19), also is located on the cytoplasmic face of the NPC and interacts physically with Nup159. Moreover, depletion of Nup82 does not affect classical NLS-mediated protein import, providing evidence for a new yeast NPC subcomplex specifically involved in RNA export.
MATERIALS AND METHODS
Plasmids and Strains.
pGEX-2TK (Pharmacia) and pGST-NUP82 (the coding region of NUP82, flanked by BamHI sites, amplified by PCR and ligated in-frame into the BamHI site of pGEX-2TK) were used to express glutathione S-transferase (GST) and a GST-Nup82 fusion protein, respectively, in bacteria. pNUP82-wt (19) and pNUP82-Δ108L [HindIII fragment of pNUP82-Δ108 (19) containing the coding region and promoter of nup82-Δ108 inserted into HindIII site of pRS315 (22)] were used to express influenza hemaglutinnin (HA)-tagged versions of wild-type Nup82 (called Nup82-wt) and a Nup82 lacking the C-terminal 108 aa (called Nup82-Δ108), respectively, under their own promoters. These two plasmids (pNUP82-wt and pNUP82-Δ108L) have the LEU2 gene as a marker. The plasmids pNUP82-wtU [HindIII fragment of pNUP82-wt (19) ligated into pRS316 (22)] and pNUP82-Δ108U [HindIII fragment of pNUP82-Δ108 (19) containing the coding region and promoter of nup82-Δ108 inserted into HindIII site of pRS316] were used like pNUP82-wt and pNUP82-Δ108L, respectively, but have the URA3 gene as a marker. pVDP7 (20), pRS315, and pGFP-URA (23) were described previously.
All yeast strains contained a disruption of the genomic copy (or copies) of NUP82 with the HIS3 gene covered with a plasmid containing either wild-type or mutant NUP82 genes described above. Yeast strains NUP82-wtU [identical to NUP82-wt (19) but containing the covering plasmid pNUP82-wtU instead of pNUP82-wt], NUP82-Δ108U [identical to NUP82-Δ108 (19) but containing pNUP82-Δ108U instead of pNUP82-Δ108], and the diploid strain NUP82-wtD [Mata/Matα his3-Δ200/his3-Δ200 leu2–3, 112/leu2–3, 112 lys2–801/lys2–801 trp1–1/trp1–1 ura3–52/ura3–52 nup82∷HIS3/nup82∷HIS3 pNUP82-wt(LEU)] were used.
Preparation of Yeast Nuclear Envelopes.
Yeast nuclear envelopes from NUP82-wtD yeast were prepared as described (24) with the following modifications. Before spheroplasting, cell walls were weakened by incubating cells at 30°C in 100 mM Tris⋅Cl, pH 9.4/10 mM DTT for 30 min and then washed twice in 1.1 M sorbitol. Digestion of cell walls was done in 1.1 M sorbitol (2.5 ml sorbitol/1 g wet weight of cells) containing 50 μl NEE-154 glusulase (NEN)/20 μl 1% Zymolyase 20-T/15 μl NovoZym 234 (Novo BioLabs, Danbury, CT; Novo Industries, Bagsvaerd, Denmark) per ml of the total solution for 3 hr at 30°C. After digestion, an equal volume of 1.1 M sorbitol was added and the cells were pelleted, washed twice in 1.1 M sorbitol, and then resuspended in 1.1 M sorbitol (5 ml/g cells). Cells were overlaid onto a cushion of 1.1 M sorbitol containing 7.5% Ficoll and pelleted and washed twice as before.
Four grams of cells were resuspended in 15 ml of a polyvinylpyrrolidone (PVP) solution (8% PVP/20 mM potassium phosphate, pH 6.5/0.75 mM MgCl2) containing 0.025% Triton X-100/0.1% solution P (87.0 mg phenylmethylsulfonyl fluoride plus 1.5 mg pepstatin A dissolved in 5 ml dry absolute ethanol) and 0.1% protease inhibitor solution (2.5 mg/ml each of leupeptin, chymostatin, and antipain) and lysed with a polytron. Solution P and the protease inhibitor solution were used in all subsequent manipulations. The lysate (Fraction 1, cell lysate) was overlaid onto a cushion of PVP solution containing 0.3 M sucrose and pelleted (Fraction 2, crude cytosol and Fraction 3, crude nuclei). The pellet (3 g) was resuspended in 2.6 ml PVP solution/2 M sucrose with the polytron. This was overlaid on a step gradient of PVP solution with 2.52, 2.25, and 2.15 M sucrose and centrifuged at 103,000 × g for 8 hr at 4°C. Fractions were collected as follows: top (Fraction 4), top/2.15 (Fraction 5), 2.15/2.25 (Fraction 6), 2.25/2.52 (Fraction 7, purified nuclei), and 2.52 (Fraction 8). Crude nuclear envelopes were prepared from Fraction 7 as described (24).
Immunoelectron Microscopy of Yeast Nuclear Envelopes.
Immunoelectron microscopy was performed as described (17). Briefly, wells of microtiter plates first were prepared by incubating with 100 μl of 2.5% glutaraldehyde (BDH) in H2O for 15–30 min at room temperature. The plates were washed in running H2O, and wells were dried by aspiration and then incubated with 100 μl of 0.1% polylysine (Sigma) for 15–30 min at room temperature. Then, polylysine was removed by aspiration and replaced with 225 μl of a 1:1 mixture of crude NE from NUP82-wtD yeast and bt-DMSO (10 mM Bis⋅Tris, pH 6.5/0.1 mM MgCl2/20% DMSO). Microtiter plates were centrifuged at 12,000 rpm in an HB4 rotor for 2 hr, and the supernatant was removed by aspiration. The NE in the wells were fixed in a solution of 4% formaldehyde (Fluka) in 1 M sucrose/bt-DMSO for 10 min at room temperature, washed twice in bt-DMSO and then once in IEM buffer (0.5× PBS/0.5%BSA/1 mM MgCl2/0.02% NaN3/10 mM CaCl2/10 mM ZnCl2/0.2% Solution P), incubated overnight at 4°C with HA.11 antibody (Berkeley Antibody, Richmond, CA) at a dilution of 1:100 in IEM buffer, washed three times in IEM buffer, incubated overnight at room temperature with 10 nm gold anti-mouse IgG (Amersham) in IEM buffer, washed twice with 0.5% PBS/1 mM MgCl2 and once with 1.25% glutaraldehyde in 0.5% PBS/1 mM MgCl2, fixed overnight at 4°C in 1.25% glutaraldehyde in 0.5% PBS/1 mM MgCl2, washed twice in 0.5% PBS/1 mM MgCl2, dehydrated with graded ethanol washes, embedded in Epon, removed from the microtiter wells, reembedded, sectioned, and visualized by transmission electron microscopy.
In Vivo Nuclear Import Assay.
The assay was done as described (23) with the following modifications. Briefly, NUP82-wt or NUP82-Δ108L cells were transformed with pNLS-GFP-URA, which constitutively expresses a green fluorescent protein (GFP) fused to the simian virus 40 Large T antigen NLS. Only freshly transformed yeast were used for the assay because cells transformed with this plasmid mutated over time. Cells were grown in medium lacking uracil (−URA) with 2% dextrose at 30°C to mid-log phase (OD600 0.2–1.0), and then half of the cells were diluted into −URA medium and incubated at 30°C for 3 more hours and the other half were diluted into −URA medium already at 37°C and grown for 3 more hours at 37°C. Cells were pelleted, washed once with sterile distilled H2O, and then resuspended in −URA medium containing 10 mM each of NaN3 and 2-deoxyglucose to inhibit ATP-dependent transport and incubated for 45 min at 30°C for the first group or at 37°C for the second group to allow equilibration of NLS-GFP throughout the cell. Then, cells were pelleted and resuspended in ice-cold sterile-distilled H2O and left on ice until use.
The assay was done by diluting cells 1:4 into −URA medium containing 2% dextrose at room temperature, placing them on a microscope slide and counting cells in a Zeiss Axiophot fluorescence microscope on the fluorescein isothiocyanate channel. Cells were scored as nuclear if a clear boundary could be seen between the nucleus and cytoplasm. Otherwise they were scored as cytoplasmic. Cells were counted up to the time point, and then the counting was started over with new cells until the next time point. At least 30 cells were counted on each time point. Each time point on a curve represents at least three separate sets of cells counted. Scoring for nuclear staining was done as described (23).
In Vitro Binding Assay.
Overlay conditions were exactly as described (25). Fusion protein was prepared as follows: pGEX-2TK or pGST-NUP82 was transformed into BLR(DE3) bacteria. Bacteria were grown in Luria–Bertani medium at 37°C until OD600 of 0.6–0.8 was reached. Protein expression was induced by addition of isopropyl β-d-thiogalactoside to 50 μM, and incubation at 37°C was continued for 1 hr. Cells were pelleted, freeze-thawed twice, and sonicated for 30 sec twice with a probe sonicator in 1/20 original culture volume of transport buffer (20 mM Hepes, pH 7.5/110 mM KOAc/2 mM MgCl2) with 1:50 of a solution of a protease inhibitor mixture (protease inhibitor tablets without EDTA, Boehringer Mannheim; 1 tablet diluted in 1 ml of H2O). This was diluted 1:1 with transport buffer/5% milk and incubated with the blot described in Fig. 3B. Rabbit polyclonal anti-GST antibody (25) was used to identify bound GST-Nup82 and GST.
Figure 3.
Nup82 binds to the coiled-coil domain of Nup159. (A) Nup159 was expressed in Escherichia coli in five different segments, p159-1, amino acid residues 1–175; p159-2, residues 176–440; p159-3, residues 441–876; p159-4, residues 877-1222; p159-5, residues 1223–1460. The position of the FXFG peptide repeats, coiled coil, and amino acid number are shown [modified from Kraemer et al. (17)]. (B) Fractions containing the expressed proteins were subjected to SDS/PAGE, transferred to nitrocellulose, incubated with GST-Nup82 or GST alone, and then incubated with a polyclonal rabbit anti-GST antibody. The arrowhead shows where p159-5 migrates. The asterisk is at approximately double the molecular weight of p159-5.
RESULTS
Depletion of Nup82 Has No Significant Effect on Nuclear Protein Import.
In an earlier study (19) it was shown that removing the carboxyl-terminal 108 aa of Nup82 rendered cells temperature-sensitive at 37°C because of rapid degradation of the mutant protein at the restrictive temperature. Nup82 was also shown to be required for poly(A)+ RNA export (18, 19), although a protein import defect was not observed in cells depleted of Nup82 (18). However, the sensitivity of the assay used did not provide conclusive evidence that Nup82 was not involved in protein import into the nucleus.
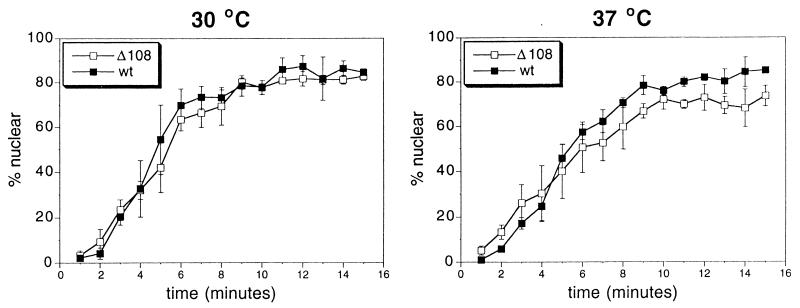
The development of a simple assay for the ability of yeast to import protein into their nuclei (23) allowed us to test more definitively whether import of protein was affected by the Nup82p-Δ108 truncation. The assay employs constitutively expressed GFP fused to the simian virus 40 Large T antigen NLS (NLS-GFP). NLS-GFP localizes to the nucleus under normal conditions. When the cell is treated with the metabolic inhibitors 2-deoxyglucose and sodium azide in the absence of glucose, active nuclear transport is blocked and, as a result, the NLS-GFP, which is under 40 kDa, is distributed equally between the cytoplasm and nucleus by diffusion. When the cells are washed and incubated with medium containing glucose, the inhibitors are diluted out and active transport begins, bringing the NLS-GFP back to the nucleus.
The assay is a time course of return of NLS-GFP to the nucleus and measures the percentage of viewed cells that show distinct nuclear staining. Two yeast strains with the genomic copies of NUP82 disrupted by HIS3 were covered with plasmids containing HA-tagged versions of either Nup82 or Nup82-Δ108 (NUP82-wt and NUP82-Δ108 cells). They were grown to mid-log phase at 30°C and then tested for ability to import or grown for an additional 3 hr at 37°C and tested. As Fig. 1 shows, no statistically significant difference in nuclear import was observed for the two strains at permissive or restrictive temperatures. The plateaus of the curves for the cells grown at the 37°C show a slight, but statistically significant difference. However, after 3 hr at the restrictive temperature, a small proportion of NUP82-Δ108 cells probably are dead, which would account for a slightly lower plateau.
Figure 1.
Depletion of Nup82 has no significant effect on nuclear protein import. NUP82-wt and NUP82-Δ108 yeast containing pGFP-URA were grown to mid-log phase and then incubated at 30°C or 37°C for 3 hr and tested in the NLS-GFP nuclear import assay.
Nup82-wt Is on the Cytoplasmic Side of the NPC.
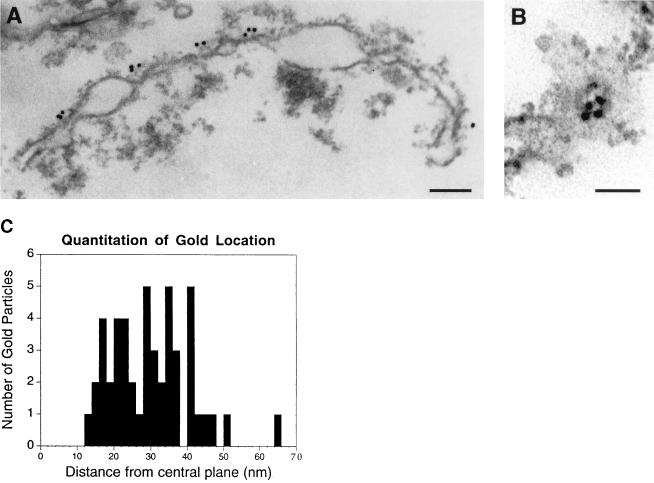
To sublocalize Nup82, immunoelectron microscopy was performed on crude NE from a diploid yeast strain with the genomic copies of NUP82 disrupted, harboring a plasmid that expresses Nup82-wt, the HA-tagged version of Nup82, under its own promoter (strain NUP82-wtD). The NE were prepared for immunoelectron microscopy and probed with mAb HA.11, which recognizes the HA epitope. Anti-HA antibody was visualized with 10 nm gold-labeled anti-mouse antibody.
All of the gold particles that were unequivocally associated with NPCs were seen only on one side of the NE in all sections observed (Figs. 2 A and B). Based on the fact that the gold was on the convex sides of the envelopes and the observation that the concave sides contain residual chromatin, we conclude that Nup82 is on the cytoplasmic side of the NPC.
Figure 2.
Immunoelectron microscopic localization of Nup82 to the cytoplasmic side of the NPC in isolated crude nuclear envelopes. (A) Side view. (Bar = 150 nm.) (B) Front view. (Bar = 55 nm.) (C) Quantitation of distance of gold particles from the NPC. The distance measured is a straight line perpendicular to the central plane of the NPC. Forty-eight particles were measured, and the mean and SD were found to be 29.9 nm and 10.9 nm, respectively.
Quantitation of the distance of the gold from the center of the NPC was performed and an average distance of 29.9 nm with an SD of 10.9 nm was found (Fig. 2C). Both the long distance from the center of the NPC and the relatively large standard deviation led to the conclusion that Nup82 is at the base of or on structures extending out from the cytoplasmic side of the NPC.
Nup82 Physically Interacts with Nup159.
Nup82 is localized to the cytoplasmic side of the NPC, and its depletion results in an export defect but lacks an import defect. Another nucleoporin, Nup159, is localized cytoplasmically (17) and gives rise to similar phenotypes upon mutation (16). Because of the similar localization and phenotype of mutant versions of the two proteins, experiments were undertaken to see whether the two proteins interact physically. Bacterial lysates containing segments of Nup159 (Fig. 3A) were separated by SDS/PAGE and then blotted onto nitrocellulose (17) and probed with bacterial lysates containing either GST or GST fused to Nup82 (GST-Nup82) at its amino terminus.
GST-Nup82 bound only to lysates expressing p159-5 (Fig. 3B) and appeared to specifically recognize a band that comigrates with the induced 159-5 band (shown by arrow) and a series of lower bands that probably represent degradation products of p159-5. 159-5 encodes the C-terminal 238 aa of Nup159 and contains a putative coiled-coil domain. A faint band can be seen consistently at twice the molecular weight of p159-5 (indicated by the asterisk), which may represent a p159-5 dimer. Notably, GST-Nup82 does not bind to segment p159-3, which contains multiple copies of the sequence FXFG and was shown previously to interact with the transport factors Kap 95 (17) and Kap 111 (25). GST alone does not recognize any bands that comigrate with the expressed segments of Nup159.
Rss1/Gle1 Partially Rescues the nup82-Δ108 ts Phenotype.
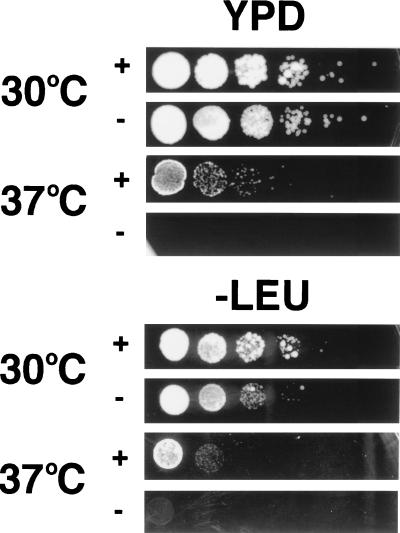
A multicopy suppressor screen of a NUP159 mutant strain (rat7-1) previously identified the protein Rss1 (20) [also identified separately as Gle1 (21)]. Overexpression of Rss1/Gle1 rescues rat7-1 cells from death at 37°C. Because of the similarity between Nup82 and Nup159, Rss1/Gle1 was analyzed for its ability to suppress the ts phenotype of NUP82-Δ108U cells. NUP82-Δ108U cells were transformed with a multicopy plasmid containing the gene encoding Rss1/Gle1 (pVDP7, generous gift of C. N. Cole and V. Del Priore, Dartmouth Medical School, Hanover, NH) or empty plasmid (pRS315). NUP82-Δ108U cells harboring either pVDP7 or pRS315 were grown overnight in selective medium, and 10-fold serial dilutions of these cultures were spotted onto plates containing either complete (YPD) or selective (-LEU) medium (Fig. 4). At the permissive temperature (30°C), the same number of colonies were observed at equivalent dilutions whether or not cells were overexpressing Rss1/Gle1, but many of the colonies of the cells overexpressing Rss1/Gle1 grew faster [compare the size of colonies containing pVDP7 (+) or pRS315 (−) after growth for the same period of time at 30°C]. At the restrictive temperature (37°C), the survival of cells containing overexpressed Rss1/Gle1 (+) is 1,000-fold higher than survival of cells that are not overexpressing Rss1/Gle1 (−). However, comparison of pVDP7-containing cells incubated at 30°C to the same cells incubated at 37°C shows that survival is much higher at 30°C, indicating that the rescue by Rss1p/Gle1p overexpression is only partial, i.e., many NUP82-Δ108U cells will not survive at the restrictive temperature (37°C) despite the overexpression of Rss1/Gle1 [which is also true for its suppression of rat7–1 cells (20)]. Consistent with the partial effect of Rss1/Gle1, growth of Rss1/Gle1 overexpressing cells on selective medium (-LEU) is significantly slower than on complete medium (YPD), presumably because of the greater metabolic requirements the cell must fulfill (in Fig. 4, compare the size of colonies grown on complete medium with those grown on selective medium at 30°C).
Figure 4.
Overexpression of Rss1/Gle1 partially rescues the ts phenotype seen in NUP82-Δ108U yeast. The (+) rows contain NUP82-Δ108U yeast harboring pVDP7 (which overexpresses Rss1/Gle1). The (−) rows contain NUP82-Δ108U yeast harboring pRS315 (empty plasmid). Cells were grown in selective medium (-LEU) at 30°C overnight. Equivalent numbers of cells and serial 10-fold dilutions (based on OD600) were plated on either complete medium (YPD) or selective medium (-LEU) and incubated at 30°C or 37°C for 4 days.
DISCUSSION
We have demonstrated that Nup82 is located on the cytoplasmic face of the NPC and physically interacts with Nup159. Localization of Nup159 showed that it is 41 nm from the center of the pore with an SD of 15 nm (17), which overlaps with the distance found for Nup82 (29.9 nm with an SD of 10.9 nm). The large SD suggests that these proteins are located on filamentous structures that are mobile during the nuclear envelope purification and electron microscopic preparation. Nup82 can be coimmunoprecipitated with Nsp1 (18), a nucleoporin located on both sides of the NPC. Nsp1’s location on both sides of the NPC has been interpreted as evidence that it is part of the symmetric core of the NPC. Nup82, therefore, may link the cytoplasmically facing structures and the NPC core.
In vertebrates, the cytoplasmic filaments emanating from the NPC are composed of Nup358 (26, 27) (which contains FXFG repeats and a Ran-binding domain), Nup214 (28, 29), and Nup88 (30) and possibly others. Nup214, the mammalian homolog of Nup159, is an FXFG repeat containing nucleoporin originally cloned as CAN (31), an oncogene identified in two types of leukemia. Subsequently, Nup88 was identified through coimmunoprecipitation with Nup214 (30). Nup88 contains a C-terminal coiled coil. Based on its domain structure, Fornerod et al. (30) suggested that it may be the functional homolog of Nup82 despite the lack of homology between the two proteins on the primary structural level. The demonstration that Nup82 and Nup159 physically interact and also localize to the cytoplasmic side of the NPC is in agreement with this idea, and we conclude that Nup159/Nup82 form part of a yeast NPC subcomplex homologous to the Nup214/Nup88 mammalian subcomplex, i.e., part of a yeast equivalent to the mammalian cytoplasmic filaments.
NUP82 and NUP159 mutations are both partially suppressed by overexpression of Rss1/Gle1. The effect of Rss1/Gle1 overexpression appears to be specific because it is not able to suppress the growth defects in mutants of the nucleoporins NUP120, NUP133, or NUP85 (20), all of which display mRNA export defects but not protein import defects. Rss1/Gle1 is required for mRNA export (20, 21), suggesting that it acts in a pathway together with Nup82 and Nup159 to export mRNA, which may account for its ability, when overexpressed, to partially suppress the effects of disruption of these proteins. Immunofluorescence microscopy of cells containing a protein A-Rss1/Gle1 fusion protein demonstrates punctate staining of the nuclear rim (21), a pattern characteristic of nucleoporins, which is consistent with Rss1/Gle1 being associated with NPCs or being an NPC component.
It is striking that both Nup82 and Nup159 are cytoplasmically located nucleoporins whose depletion inhibits mRNA export without affecting classical NLS-mediated import. Presently, no model explains this result. Perhaps export is more sensitive to certain NPC perturbations than import or the cytoplasmic components of the NPC have a special role promoting movement of export substrate away from the exit of the NPC. Resolution of these issues will depend on further understanding of the import and export machinery in addition to that of the structure of the NPC.
Acknowledgments
We thank Helen Shio at the Rockefeller University Electron Microscopy Facility for assistance in performing electron microscopic studies. We also thank Lucy Pemberton, Jonathan Rosenblum, and Roland Beckmann for critical reading of the manuscript, Charles N. Cole and Valerie Del Priore for pVDP7, and David Goldfarb for pGFP-URA. M.E.H. was supported by National Institutes of Health Medical Scientist Training Program Grant GM07739 to the Cornell/Rockefeller/Sloan-Kettering Tri-institutional M.D.-Ph.D. program.
ABBREVIATIONS
- Nup
nucleoporin
- NPC
nuclear pore complex
- NE
nuclear envelope
- HA
hemagglutinin
- NES
nuclear export sequence
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- NLS
nuclear localization sequence
References
- 1.Reichelt R, Holzenburg A, Buhle E J, Jarnik M, Engel A, Aebi U. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Q, Rout M P, Akey C W. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 3.Rout M P, Wente S R. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 4.Akey C W, Radermacher M. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unwin P N, Milligan R A. J Cell Biol. 1982;93:63–75. doi: 10.1083/jcb.93.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarnik M, Aebi U. J Struct Biol. 1991;107:291–308. doi: 10.1016/1047-8477(91)90054-z. [DOI] [PubMed] [Google Scholar]
- 7.Ris H. EMSA Bull. 1991;21:54–56. [Google Scholar]
- 8.Ris H. Application of Low Voltage High Resolution SEM in the Study of Complex Intracellular Structures, Proceedings of the XII International Congress on Electron Microscopy. San Francisco: San Francisco Press; 1990. [Google Scholar]
- 9.Goldberg M W, Allen T D. J Cell Biol. 1992;119:1429–1440. doi: 10.1083/jcb.119.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurt E C, Mutvei A, Carmo-Fonseca M. Intl Rev Cytol. 1992;136:145–184. doi: 10.1016/s0074-7696(08)62052-5. [DOI] [PubMed] [Google Scholar]
- 11.Wente S R, Rout M P, Blobel G. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wimmer C, Doye V, Grandi P, Nehrbass U, Hurt E C. EMBO J. 1992;11:5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grandi P, Schlaich N, Tekotte H, Hurt E C. EMBO J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nehrbass U, Rout M P, Maguire S, Blobel G, Wozniak R W. J Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aitchison J D, Rout M P, Marelli M, Blobel G, Wozniak R. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorsch L C, Dockendorff T C, Cole C N. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraemer D M, Strambio-de-Castillia C, Blobel G, Rout M P. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- 18.Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt E C. J Cell Biol. 1995;130:1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurwitz M E, Blobel G. J Cell Biol. 1995;130:1275–1281. doi: 10.1083/jcb.130.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Priore V, Snay C A, Bahr A, Cole C N. Mol Biol Cell. 1996;7:1601–1621. doi: 10.1091/mbc.7.10.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy R, Wente S R. Nature (London) 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 22.Sikorski R S, Heiter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shulga N, Roberts P, Gu Z, Spitz L, Tabb M M, Nomura M, Goldfarb D S. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strambio-de-Castillia C, Blobel G, Rout M P. J Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pemberton L F, Rosenblum J S, Blobel G. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, Matunis M J, Kraemer K, Blobel G, Coutavas E. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 27.Delphin C, Guan T, Melchior F, Gerace L. Mol Biol Cell. 1997;8:2379–2390. doi: 10.1091/mbc.8.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraemer D, Wozniak R W, Blobel G, Radu A. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Deursen J, Boer J, Kasper L, Grosveld G. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- 30.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Gopal Murti K, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]