Abstract
G2 was defined originally as the temporal gap between the termination of DNA replication and the beginning of mitosis. In human cells, the G2 period was estimated to be 3–4 h. However, the absence of replicative DNA synthesis during this period designated G2 has never been shown conclusively. In this report, we show that, at some autosomal and X linked loci, programmed DNA replication continues within 90 min of mitosis. Furthermore, the major accumulation of cyclin B1, a cell-cycle marker that is usually ascribed to G2, overlaps extensively with very late DNA replication. We conclude that the G2 period is much shorter than previously thought and may, in some cells, be nonexistent.
The eukaryotic cell cycle consists of an orderly series of events in which cells grow, replicate their DNA, and segregate the duplicated genome to the daughter cells. Our current textbook view of the cell cycle was first put forth by Howard and Pelc (1) as encompassing four phases: S phase, the time in which replicative DNA synthesis occurs; M phase, the time in which mitotic chromosome segregation and cell division proceed; and two gap phases, G1 and G2, that temporally separate S phase from mitosis.
The G2 phase was inferred from the length of time between the addition of radiolabeled DNA precursors and the appearance of labeled, condensed mitotic chromosomes (1). Autoradiography, however, may not be sufficiently sensitive to detect low levels of replicative synthesis in the G2 phase. The absence of detectable incorporation of radiolabel could thus be taken to indicate erroneously the absence of DNA replication. Furthermore, DNA repair synthesis can occur throughout the cell cycle (2, 3); replicative and repair DNA synthesis are not easily distinguishable by the autoradiographic method. An additional problem is that no cellular marker signaling the termination of replicative DNA synthesis has been identified. These features contribute to an inherent lack of both sensitivity and specificity for accurately defining the terminal border of S phase.
Our studies on fragile sites and the replication timing of FMR1, the gene responsible for the fragile X syndrome, first brought our attention to the possibility of replication very close to mitosis (4). In cell lines derived from patients with fragile X syndrome, the replication of a large region surrounding the FMR1 gene is abnormally late, occurring within the G2/M compartment defined by DNA content (5, 6). Surprisingly, equally late replication was observed for several loci on the inactive X chromosome in normal female fibroblasts (7, 8), as well as on the inactive X chromosome in human–hamster hybrids (8). In this report, we have determined more precisely the timing of late DNA replication in the human cell cycle, by using a new, sensitive method for fractionating cells late in the cell cycle (9). We address two questions. Does this very late replication occur during what is usually termed G2? What proportion of the genome replicates in this late compartment?
MATERIALS AND METHODS
Cell Culture.
Lymphoblastoid cell lines were grown as described (9). Cell lines used in this study included: FF, derived from a normal male (5); VK and SK, derived from normal females; and 5106, derived from a phenotypically normal male (kindly provided by B. A. Oostra, Erasmus University, Rotterdam, Netherlands, and described below). Fresh blood lymphocytes were taken from two normal females, isolated by centrifugation over a Ficoll gradient, and cultured at 5 × 106 cells per ml in RPMI medium 1640 plus 10% fetal bovine serum. Cells were stimulated to divide with the addition of 20 μl/ml phytohemagglutanin for 3 days before BrdUrd labeling.
General PCR Conditions.
All PCRs contained 18.2 μl of PCR Supermix (GIBCO/BRL), each primer at 2 μM, and 1 μl of the BrdUrd-labeled DNA isolated from each fraction. Parameters for all PCRs included denaturation at 95°C for 5 min and 23 cycles that included denaturation at 95°C for 1 min, annealing at a temperature described below for each primer pair for 2 min, and extension at 72°C for 3 min. A final extension was carried out at 72°C for 7 min. PCR products were run on 1.2% Tris-base, acetic acid, EDTA (TAE) agarose gels. Transfer of DNA to Hybond N+ membrane filters was carried out with 0.4 M NaOH, followed by neutralization with 0.4 M Na2HPO4. Hybridization was carried out at 65°C in FBI solution (10% polyethylene glycol 8000/7% SDS/1.5× standard saline phosphate/EDTA/100 μg/ml sheared salmon sperm DNA) for at least 3 h. Thereafter, filters were washed twice in 3× standard saline citrate with 1% SDS, then washed twice in 1× standard saline citrate with 1% SDS, and exposed to x-ray film.
The following annealing temperatures were used: G6PD at 50°C, F9 at 60°C, PGK1 at 62°C, FMR1 at 60°C, inter-Alu 6–84 at 45°C, inter-Alu 6–82 at 45°C, inter-Alu H-1 at 40°C, and inter-Alu H-8 at 40°C. The oligonucleotide primer pairs used for PCR of G6PD, F9, and PGK1 have been described (5). The primer pair and corresponding size used for FMR1 are as follows: fmr69 (5′-gagtcaagtggggaagactaagttgc-3′) and fmr357 (5′-gaactttgtgcataccctctgcttcc-3′) (291 bp). The oligonucleotide primer pairs employed for allele-specific inter-Alu PCR and the corresponding sizes are as follows: inter-Alu 6–84, 6–84F (5′-agtatccacaggtatctga-3′) and 6–84R (5′-ttagaaagtacattaggcac-3′) (240 bp); inter-Alu 6–82, 6–82A (5′-cgatatgaagaaaataaaag-3′) and 6–82B (5′-atctttaatctcatctccat-3′) (246 bp); inter-Alu H-1, H-1F (5′atcccatacatagacca-3′) and H-1R (5′-agtaaagcattgatcca-3′) (262 bp); inter-Alu H-8, H-8F (5′-tagcatttgattttcta-3′) and H-8R (5′-ttctctattattacgga-3′) (225 bp).
Replication Timing with Cyclin B1 Flow Cytometry.
Flow cytometry was performed as described (9), with a few exceptions. DNA content was measured by resuspending the cells in 1 ml of the staining solution as described (5). In some experiments, 20 μl of antibody (FITC-conjugated Cyclin B1 Antibody Reagent Set; PharMingen) was used instead of 0.5 μg of the antibody with no discernible differences in results.
Previously, we have described a method for studying replication timing that uses the incorporation of BrdUrd (5). We used this method with the following changes: the immunoprecipitation of BrdUrd-labeled DNA was carried out with 1 μg of anti-BrdUrd monoclonal antibody and 16.5 μg of rabbit IgG directed against mouse IgG; lyophilized BrdUrd DNA isolated from each sorting fraction was dissolved in a volume of Tris-EDTA to obtain the equivalent of 400 cells per μl; and BrdUrd-labeled CHO-YH21 DNA was not added. All cell fractionation isolates were controlled internally by assaying characteristic early and late replicating sequences (for example, see H-8, Fig. 3C). Individual sequence profiles were obtained in at least two different cell lines.
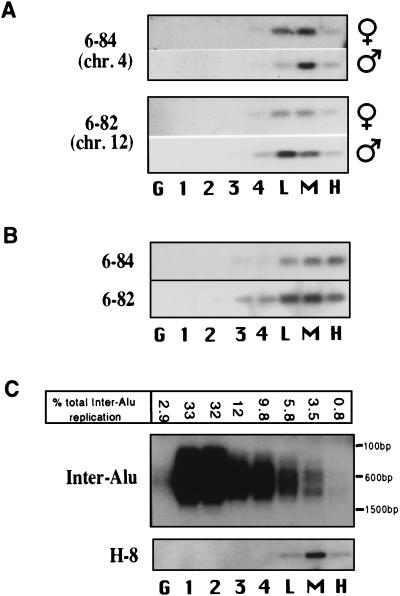
Figure 3.
Detection of very late DNA replication for autosomal loci. (A) Replication of two inter-Alu sequences located on chromosomes 4 and 12 is highly restricted and very late in the cell cycle in lymphoblasts. The high cyclin B1 fraction contains on average 13% of the replication signal for these loci. (B) In phytohemagglutinin-stimulated lymphocytes, replication of the same autosomal inter-Alu sequences described in A is also highly restricted and very late in the cell cycle. In this case, the high cyclin B1 fraction contains 26% of the replication signal for these autosomal sequences. (C) Replication timing for a heterogeneous population of inter-Alu sequences is spread throughout the cell cycle in lymphoblasts, with about 1% occurring within the high cyclin B1 fraction. The percentage of the total inter-Alu replication detected by phosphorimager analysis is shown for each cell-cycle fraction. The same membrane was stripped of the labeled primer and reprobed with a specific late replicating inter-Alu sequence found on the X chromosome (H-8); the results demonstrated the specificity of this approach. An alternative quantification method in which radioactive deoxyribonucleoside triphosphates were incorporated during the PCR gave similar results.
Quantitation of the hybridization signals was performed by densitometry analysis. To calculate the percent of DNA replication in the mid- and high cyclin B1 fractions for loci on the inactive X chromosome, we assumed that the total signal constituted the later replication signal. In profiles for which both phosphorimager analysis and densitometry data were available, the calculated percent of total DNA replication was highly comparable.
Isolation and Analysis of Autosomal Late Replicating Sequences.
We isolated a number of sequences by PCR amplification between closely spaced Alu repeats by using the primer Alu 3.2 (5′-gcgacagagcgagactccgtctc-3′). This primer is specific to the 3′ end of the Alu consensus sequences that represent five of the eight currently defined subclasses (10). BrdUrd DNA isolated from either the G2/M cell-cycle fraction or the high cyclin B1 fraction was used to amplify late replicating inter-Alu sequences. PCR conditions were as follows: denaturation at 95°C for 5 min; 30 cycles that included denaturation at 95°C for 1 min, 65°C for 2 min, and 76°C for 3 min; and a final extension of 76°C for 5 min. PCR products were then cloned using the TA Cloning Kit (Invitrogen). Individual clones that were confirmed as replicating late in the cell cycle were then used for further analysis. The chromosomal location of each sequence was determined either by probing inter-Alu products amplified from a human-rodent somatic cell mapping panel (Coriell Cell Repositories, Camden, NJ) or by directly performing the locus-specific PCR on the same hybrid DNA. Inter-Alu clones 6–84, 6–82, H-1, and H-8 are located on chromosomes 4, 12, 10, and X, respectively.
Quantification of Generalized Inter-Alu PCR.
One microliter of BrdUrd DNA equal to 400 cell equivalents from each fraction was amplified by using the Alu3.2 primer as described above, except that the products were amplified with 28 cycles. Then, the products were separated electrophoretically and transferred to a Hybond N+ membrane. The blot was then hybridized at 42°C overnight in FBI hybridization solution. The blot was washed with 6× standard saline citrate and 1% SDS at 42°C for 60 min and either exposed to x-ray film or quantified by phosphorimager analysis. The percentage of inter-Alu replication detected in each fraction was determined by dividing the measured signal intensity in each lane by the total measured intensity.
An alternative quantitative approach incorporated radionucleoside triphosphates. PCR was performed with 200 μM dGTP, 200 μM dATP, 100 μM dCTP, 100 μM TTP, 10 μCi of [32P]dCTP, 10 μCi [32P]TTP, 4 μM Alu3.2, 1.25 units Taq polymerase, and 1 μl of BrdUrd DNA in a 20-μl reaction by using a standard buffer (Boehringer Mannheim). The inter-Alu PCR was run at the conditions described above, except that 23 cycles were performed. Products were separated electrophoretically on a 1.3% TAE agarose gel and transferred to a Hybond N+ membrane. Incorporation of radiolabeled nucleotides into the PCR products amplified with BrdUrd DNA from each replication-timing fraction was quantified by phosphorimager analysis. The percentage of inter-Alu replication ongoing in each cell-cycle fraction was calculated as described above, yielding percentages of inter-Alu replication of 1.3, 29, 33, 18, 9.8, 4.8, 2.9, 0.8 for G1 through high cyclin B1, respectively.
The cell line 5106 (used to obtain the data shown in Fig. 3B) was derived from a normal male with a large expansion of the CGG repeat in the FMR1 gene but without associated methylation (kindly provided by B. A. Oostra). We did not detect any differences in the replication-timing profiles of several specific inter-Alu sequences between this and other cell lines, confirming that the expansion of the CGG repeat does not have a general effect on replication timing (5). In addition, comparable results were obtained by using the normal male cell line, FF.
RESULTS
Cell-Cycle Fractionation for Replication Timing.
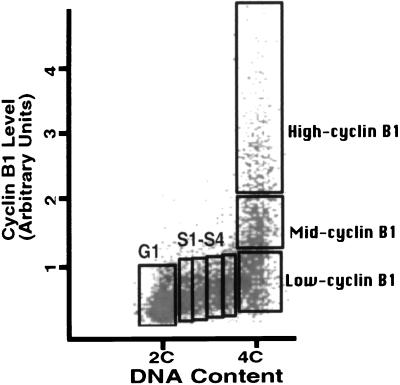
To investigate very late DNA replication, we have developed a method using cyclin B1 to identify cells at different times within the late periods of the cell cycle (9). Levels of this cell-cycle protein rise abruptly as cells obtain a 4C DNA content, peak during the metaphase/anaphase transition, and fall precipitously as cells complete mitosis (11). The increased level of cyclin B1 is thus a useful marker for cells very late in the cell cycle; this increase in cyclin B1 has been considered a marker for G2 and the early stages of mitosis (12, 13). Our flow cytometry method takes advantage of this accumulation of cyclin B1 to sort cell populations progressively closer to the mitotic transition. We analyzed eight cell-cycle fractions isolated on this basis: the G1 phase fraction, four S phase fractions that were clearly between the 2C and 4C peaks (S1–S4) and three fractions with a DNA content of about 4C with low, mid-, and high levels of cyclin B1 (Fig. 1). We previously estimated that 4C cells with low and mid-levels of cyclin B1 transit to high levels of cyclin B1 in about 3.0 and 1.6 h, respectively (9).
Figure 1.
Cell-cycle fractionation by cyclin B1 flow cytometry. Human lymphoblastoid cells were labeled with BrdUrd for 90 min (5), fixed, and separated on the basis of DNA content and cyclin B1 levels. The cyclin B1 units are arbitrary. Equal numbers of cells (between 5,000 and 10,000) were collected from each sorting fraction (G1; S1–S4; and low, mid-, and high cyclin B1) and used for subsequent analysis of replication timing (modified from ref. 9).
Replication Timing of X Linked Loci.
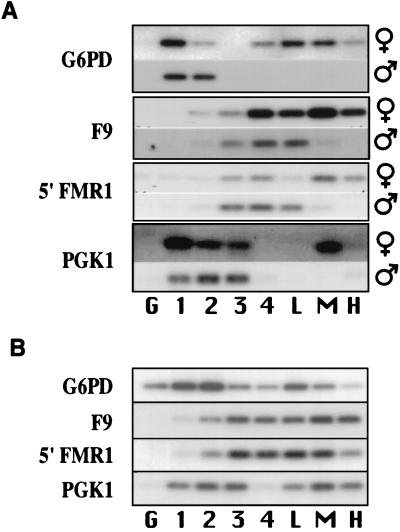
We coupled our cyclin B1 method of fractionating cells and our 5′ bromodeoxyuridine (BrdUrd) method to isolate newly replicated DNA (5) to examine the replication timing for four X linked loci; glucose-6-phosphate dehydrogenase (G6PD; Xq28), blood factor IX (F9; Xq26.3), fragile X mental retardation 1 (FMR1; Xq27.3), and phosphoglycerate kinase 1 (PGK1; Xq13.3). In normal male lymphoblasts, the X linked loci analyzed have profiles representative of the replication timing for these regions on an active X chromosome (Fig. 2A). In normal female lymphoblasts, a bimodal replication pattern is seen for each locus, with the later replication signal representing the replication timing of the locus on the inactive X chromosome (7, 8). The high cyclin B1 fraction contains on average 24% (range: 18–31%) of the replication signal for loci on the inactive X chromosome. The frequent detection of a locus in cells recovered from the high cyclin B1 fraction indicates that the replication of this sequence commonly overlaps the major increase in cyclin B1. In addition, we have observed previously that about 50% of the cells in the high cyclin B1 fraction are mitotic (9). Given this observation, and the 90 min BrdUrd-labeling period, we estimate that DNA replication detected in cells recovered from the high cyclin B1 fraction must have occurred on average within 90 min of mitosis. More generally, the average sum of the inactive X replication signals in the mid- and high cyclin B1 fractions is 71% (range: 48–84%); thus, over two-thirds of the cells have replicated these loci within 3 h of mitosis.
Figure 2.
Detection of very late DNA replication for loci on the inactive X chromosome. (A) Newly replicated DNA at four loci on the inactive X chromosome is detected in lymphoblasts recovered from all three cyclin B1 fractions. Replication-timing profiles for several X linked loci in male and female cell lines are shown. The high cyclin B1 fraction contains on average 24% of the replication signal for loci on the inactive X chromosome. (B) In phytohemagglutinin-stimulated lymphocytes, newly replicated DNA at four loci on the inactive X chromosome is detected in cells recovered from all three cyclin B1 fractions. The high cyclin B1 fraction contains on average 22% of the replication signal for loci on the inactive X chromosome. The detection of newly replicated DNA in the high cyclin B1 fraction demonstrates that the replication of sequences close to mitosis in normal human cells is not a consequence of cell transformation.
Replication Timing of Autosomal Loci.
Does replication of sequences other than those on the inactive X chromosome also extend into this very late part of the cell cycle? We characterized the replication timing of several autosomal loci that were selected for replicating late in the cell cycle (see Materials and Methods). Profiles from four late replicating autosomal loci show that replication had occurred in cells subsequently collected in all three cyclin B1 fractions, including the high cyclin B1 fraction (Fig. 3A; data not shown for the two additional loci). The high cyclin B1 fraction contains on average 13% (range: 7–19%) of the replication signal for these autosomal sequences. This result indicates that the DNA replication of several autosomal sequences overlaps with the accumulation of cyclin B1 in some cells, similar to loci examined on the inactive X chromosome. Furthermore, the detection of replication in cells recovered from the high cyclin B1 fraction indicates that in some cells autosomal loci replicate within 90 min of entry into mitosis and during the major increase in cyclin B1. Furthermore, the average sum of the replication signals in the mid- and high cyclin B1 fractions is 56% (range: 42–80%), indicating that over half of the cells have replicated these autosomal sequences within 3 h of mitosis. As expected for most autosomal loci, we did not detect a significant difference in the replication profiles between the male and female cell lines. The phenomenon of very late replication is therefore not restricted to loci on the inactive X chromosome.
Generalized Replication Timing.
What fraction of total genomic replication can we assign to this very late replicating component? We examined the replication timing of a large subset of genomic sequences by using a primer designed to amplify sequences found between closely spaced Alu sequences; we refer to these as inter-Alu sequences. Limited-cycle PCR was used to amplify a heterogeneous population of inter-Alu sequences found within each replication-timing fraction. Quantification of the PCR products provides an estimate of the amount of inter-Alu replication that was ongoing in each cell-cycle fraction. Inter-Alu replication is skewed toward the first half of S phase (Fig. 3C), in agreement with previous studies (14). About 5%, 3%, and 1% of the total inter-Alu DNA replication are detected in cell fractions having low, mid-, and high cyclin B1 levels, respectively. The detectable DNA replication for these sequences in the high cyclin B1 fraction suggests that an appreciable fraction of the genome—about 1%—is replicated on average within 90 min of mitosis; the sum of the replication detected in the mid- and high cyclin B1 fractions indicates that at least 4% replicate within 3 h of mitosis. Given the overrepresentation of the inter-Alu replication in early S phase, these values for very late DNA replication may be underestimated.
Replication Timing in Nontransformed Cells.
To determine whether DNA replication close to mitosis also occurs in cells that had not been transformed, we studied the replication timing of several loci in phytohemagglutinin-stimulated lymphocytes. In such cells, G2 has been estimated to be 3–3.5 h (15–17). Very late replication, detectable in the high cyclin B1 fraction, was observed for both X linked (Fig. 2B) and autosomal sequences (Fig. 3B). The high cyclin B1 fraction contains on average 22% (range: 6–33%) and 26% (range: 20–32%) of the replication signal for inactive X linked and autosomal sequences, respectively. Replication of sequences very late in the cell cycle, overlapping extensively with the major accumulation of cyclin B1, can therefore occur in normal human cells and is not a consequence of cell transformation.
DISCUSSION
We have discovered that DNA replication overlaps extensively with the major increase in cyclin B1, previously described as a molecular marker for G2 and the early stages of mitosis (12, 13). Very late DNA replication continues within 90 min of mitosis in some cells, includes loci on the inactive X chromosome in female cells as well as autosomal loci in both female and male cells, and accounts for at least 1% of the total DNA. We have observed this phenomenon in both fresh blood lymphocytes stimulated with phytohemagglutinin and in lymphoblasts transformed with Epstein–Barr Virus.
We conclude that our observations reflect true replicative synthesis late in the cell cycle rather than repair synthesis. For a particular autosomal late replicating sequence, we can assign at least part of the detected signal to replicative DNA synthesis, because no additional major signal is seen earlier in the cell cycle (Fig. 3A and 3B). In addition, we detect a second, later component of DNA replication of X linked loci only in female cells (Fig. 2A). The absence of these later replication signals for X linked loci in male cells indicates that very late replication detected in female cells for these loci is not due to specific repair replication.
Originally, the length of G2 was estimated autoradiographically by determining the time at which 50% of metaphases were labeled after the addition of radioactive DNA precursors (18). Examination of the data in many of these early publications (15–17, 19) reveals that a small percentage of labeled metaphases was seen commonly after 1- to 2-h periods of labeling, a considerably shorter time than the 3–4 h estimated for G2. These data indicate that the length of G2 in some cells may be markedly shorter than previously estimated.
Technical limitations of previous methods made the analysis of late replicating sequences difficult. In addition, the realization that repair synthesis can occur throughout the cell cycle (2, 3), most notably in G2, later provided a possible biological explanation for the observed G2 labeling, and perhaps distracted attention from the possibility of bona fide replicative synthesis late in the cell cycle. The discovery of and emphasis on cell-cycle checkpoints bolstered the concept of a discrete G2 phase and its role as a important compartment in the cell cycle. The concept of the cell cycle consequently evolved to include a discrete G2 phase defined as the period absent of DNA replication before mitosis (Fig. 4A) (20–23), even though early studies suggested that some DNA replication might occur very late in the cell cycle. The findings presented in this paper demonstrate that programmed replication of specific sequences can account for this very late replication. We have defined more precisely the timing of this very late replication, identified some of the sequences involved, and demonstrated that a substantial fraction of cells are experiencing very late DNA replication.
Figure 4.
The cell cycle. (A) The textbook view: the cell cycle is divided into four phases; G1, S phase, G2, and mitosis. (B) A more recent view: the accumulation of cyclin B1 identifies a cell as residing in G2 or early mitosis. (C) Our proposed revision: low levels of replicative DNA synthesis (shading) are ongoing much later in the cell cycle than previously suspected, overlapping extensively with the accumulation of cyclin B1. In some cells, the existence of a discrete G2 phase is questioned.
The major increase in the levels of cyclin B1 has been described as a marker for cells in G2 (Fig. 4B) (12, 13). We have shown that in a substantial fraction of cells, the replication of several specific loci extensively overlaps with the major accumulation of cyclin B1. This result demonstrates that DNA replication is able to proceed concurrently with this late cell-cycle event. We propose an alternative description of the cell cycle in which the G2 period is very short or perhaps nonexistent for some cells and in which replicative DNA synthesis continues during the major increase in cyclin B1 (Fig. 4C).
What are the implications of ongoing DNA replication late in the cell cycle? A checkpoint mechanism involved in coupling mitosis to the completion of DNA replication has been described (for a review, see ref. 24). In response to very late DNA replication, this surveillance mechanism would ensure that mitosis is not initiated until the genome has been replicated fully. There is evidence, however, that a threshold level of unreplicated DNA may be required to trigger a cell-cycle delay (25), suggesting that low levels of DNA replication late in the cell cycle may not be sufficient to delay mitotic entry. Perhaps an additional checkpoint may function during prophase or metaphase to ensure that DNA replication is completed fully before anaphase. The absence or occasional failure of such a mechanism would lead to the anaphase separation of an incompletely replicated chromosome, with subsequent chromosomal breakage and aberrations (4). Sequences that replicate very late would also have a shortened period of time for DNA repair before cell division, possibly leading to inefficient repair and higher mutation rates. These factors can put the cell at a greater risk for genomic instability and human disease (26, 27).
In most situations, this very late DNA replication may have no adverse effect on normal cell function. The nature of the chromatin of very late replicating sequences may be such that a lengthy period of time for condensation between replication and mitosis is not required. The hypercondensed, inactive X chromosome offers a possible example of such a phenomenon.
Very late DNA replication may also play a positive role in normal cell function. The identification of sequences that specifically replicate very late in the cell cycle suggests that an active mechanism is involved in the partitioning of these loci to a very late replicating compartment. This process may be involved in the transcriptional inactivation of some sequences and their maintenance in the inactive state (8, 28). Additionally, the timing of the replication of such sequences may serve as a cellular signal for entry into mitosis.
Acknowledgments
We thank Andrew Berger and Michelle Black for excellent assistance with the flow cytometry; Theresa Canfield and Kevin Henne for technical assistance; Tina Kajimura and Jennifer Stoeck for important contributions to this work; Brian Reid and members of the Laird Lab for helpful discussions; and Lester Goldstein, Lee Hartwell, Eric Foss, and Peter Rabinovich for helpful comments on the manuscript. This work was supported by grants from the National Institutes of Health (GM53805, P30CA1570422, GM07266, GM52463, and HD16659).
References
- 1.Howard A, Pelc S R. Heredity. 1953;6:261–273. [Google Scholar]
- 2.Rasmussen R E, Painter R B. J Cell Biol. 1966;29:11–19. doi: 10.1083/jcb.29.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giulotto E, Mottura A, Carli L D, Nuzzo F. Exp Cell Res. 1978;113:415–420. doi: 10.1016/0014-4827(78)90383-x. [DOI] [PubMed] [Google Scholar]
- 4.Laird C, Jaffe E, Karpen G, Lamb M, Nelson R. Trends Genet. 1987;3:274–281. [Google Scholar]
- 5.Hansen R S, Canfield T K, Lamb M M, Gartler S M, Laird C D. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 6.Hansen R S, Canfield T K, Fjeld A D, Mumm S, Laird C D, Gartler S M. Proc Natl Acad Sci USA. 1997;94:4587–4592. doi: 10.1073/pnas.94.9.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawame H, Gartler S M, Hansen R S. Hum Mol Genet. 1995;4:2287–2293. doi: 10.1093/hmg/4.12.2287. [DOI] [PubMed] [Google Scholar]
- 8.Hansen R S, Canfield T K, Fjeld A D, Gartler S M. Hum Mol Genet. 1996;5:1345–1353. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- 9.Widrow R J, Rabinovitch P S, Cho K, Laird C D. Cytometry. 1997;27:250–254. doi: 10.1002/(sici)1097-0320(19970301)27:3<250::aid-cyto6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 10.Claverie J M, Makalowski W. Nature (London) 1994;371:752. doi: 10.1038/371752a0. [DOI] [PubMed] [Google Scholar]
- 11.Pines J, Hunter T. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pines J, Hunter T. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 13.Sherwood S W, Rush D F, Kung A L, Schimke R T. Exp Cell Res. 1994;211:275–281. doi: 10.1006/excr.1994.1087. [DOI] [PubMed] [Google Scholar]
- 14.Ten Hagen K G, Gilbert D M, Willard H F, Cohen S N. Mol Cell Biol. 1990;10:6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cave M D. J Cell Biol. 1966;29:209–222. doi: 10.1083/jcb.29.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki M S, Norman A. Nature (London) 1966;210:913–914. doi: 10.1038/210913a0. [DOI] [PubMed] [Google Scholar]
- 17.Gavosto F, Pegoraro L, Masera P, Rovera G. Exp Cell Res. 1968;49:340–358. doi: 10.1016/0014-4827(68)90185-7. [DOI] [PubMed] [Google Scholar]
- 18.Baserga R, Wiebel F. Int Rev Exp Pathol. 1969;7:1–30. [PubMed] [Google Scholar]
- 19.Zielke H R, Littlefield J W. Methods Cell Biol. 1974;8:107–121. doi: 10.1016/s0091-679x(08)60447-1. [DOI] [PubMed] [Google Scholar]
- 20.Cambell N A. Biology. Redwood City, CA: Benjamin/Cummings; 1993. p. 207. [Google Scholar]
- 21.Murray A, Hunt T. The Cell Cycle: An Introduction. New York: Oxford University Press; 1993. p. 9. [Google Scholar]
- 22.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular Biology of the Cell. New York: Garland; 1994. p. 865. [Google Scholar]
- 23.Lodish H, Baltimore D, Berk A, Zipursky S L, Matsudaira P, Darnell J. Molecular Cell Biology. New York: Scientific American Books; 1995. p. 148. [Google Scholar]
- 24.Nurse P. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 25.Smythe C, Newport J W. Cell. 1992;68:787–797. doi: 10.1016/0092-8674(92)90153-4. [DOI] [PubMed] [Google Scholar]
- 26.Weinert T, Lydall D. Semin Cancer Biol. 1993;4:129–140. [PubMed] [Google Scholar]
- 27.Hartwell L H, Kastan M B. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 28.Goldman M A, Holmquist G P, Gray M C, Caston L A, Nag A. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]