Abstract
LMO4 is a novel member of the LIM-only (LMO) subfamily of LIM domain-containing transcription factors. LMO1, LMO2, and LMO4 have distinct expression patterns in adult tissue, and we demonstrate that nuclear retention of LMO proteins is enhanced by the nuclear LIM interactor (NLI). In situ hybridization to early mouse embryos of 8–14.5 days revealed a complex pattern of LMO4 expression spatially overlapping with NLI and LHX genes. LMO4 expression in somite is repressed in mice mutant for the segment polarity gene Mesp2 and expanded in Splotch mutants. During jaw and limb outgrowth, LMO4 and LMO2 expression define mesenchyme that is uncommitted to regional fates. Although both LMO2 and LMO4 are activated in thymic blast cells, only LMO4 is expressed in mature T cells. Mesenchymal and thymic blast cell expression patterns of LMO4 and LMO2 are consistent with the suggestion that LMO genes inhibit differentiation.
Keywords: LHX genes/nuclear LIM interactor/cranial neural crest/Schwann cells/somite
The LIM domain, an approximately 55-residue, cysteine-rich zinc-binding motif, is present in a variety of proteins including LIM homeobox (LHX) proteins that contain two LIM domains and one homeodomain. LHX genes are expressed in many types of neurons and other cell types, and deletion of LHX genes results in the loss of cell fate (1). Mice mutant for LHX1 have diminished organizer activity that results in lack of head structures anterior to rhombomere 3 (2). In the central nervous system, development of forebrain and pituitary derivatives are defective in mice mutant for LHX2, LHX3, or LHX4 (1), while activation of the LHX gene Isl1 is essential for the survival of motor neurons and neighboring interneurons (3).
LMO2 represents a family of nuclear LIM-only (LMO) proteins that lack a DNA-binding homeodomain (4, 5). Unregulated LMO2 expression induces T cell tumors (6), while deletion blocks hematopoietic development (7, 8). The mechanism of LMO2 activity is thought to be the LIM domain-dependent assembly of transcription complexes and transcription regulation (9).
LIM domains of nuclear proteins bind with high affinity to the widely expressed nuclear LIM interactor (NLI) and with lesser affinity to other transcription factors (10–12). Dimeric NLI supports assembly of heteromeric complexes of LIM proteins (13), and CHIP, the Drosophila ortholog of NLI, mediates enhancer–promoter interactions of the cut and ultrabithorax genes, presumably by complex formation with transcription factors (14).
To identify novel LIM domain transcription factors, we screened two mouse embryonic expression libraries by using the LIM interaction domain (LID) of NLI. We report the isolation and characterization of LMO4, a novel LIM-only gene, which is highly expressed in the T lymphocyte lineage, cranial neural crest cells, somite, dorsal limb bud mesenchyme, motor neurons, and Schwann cell progenitors. Somitic expression of LMO4 is repressed in mice mutant for the segment polarity gene Mesp2. LMO4 and LMO2 expression in the jaw, limb, and thymus defines cells that are uncommitted to cell fates. Interaction with NLI mediates the nuclear retention of LMO proteins that lack a nuclear localization sequence.
MATERIALS AND METHODS
Expression Library Screening and Sequence Analysis.
A cDNA fragment encoding the LID (NLI amino acids 300–338) was amplified by PCR with Pfu polymerase (Stratagene) and subcloned into pGEX-2TK (Pharmacia). The GST-TK-LID fusion protein was purified by standard procedures, labeled with [γ-32P]ATP, and used to screen mouse E12 and E16 λ-ExLox expression libraries (Novagen) as described previously (10). Positive clones were purified and the cDNAs were subcloned from phage DNA. Sequence information from the 5′ end of each clone was analyzed by blast (http://www.ncbi.nlm.nih.gov/BLAST/). Phylogenetic sequence analysis was performed by using the pileup, distances, and growtree programs of the GCG software package.
RNA Purification and PCR Analysis.
Total RNA from flow cytometry-sorted thymocytes was isolated by using Rneasy spin columns (Qiagen). For reverse transcription–PCR, 200 ng total RNA from each thymic subset was converted to cDNA by using the Superscript II kit (GIBCO/BRL), and resulting samples were subjected to 35 cycles of the PCR by using Boehringer Taq polymerase and 7.5 pmol of each primer. Amplified material was resolved by agarose gel electrophoresis and visualized with ethidium bromide.
In Situ Hybridization and Immunohistochemistry.
In situ hybridization of [35S]UTP- or digoxygenin-labeled probes to tissue sections was performed as described previously (15–17). Linear templates for probe synthesis were generated as follows: LMO4pBSIIKS+/AccI for 750-bp probe, LMO2pGEM3z/EcoRI for 1.1-kb probe; LHX3pcDNA3/EcoRI; SOX10pZL1/AvaI for 1-kb probe; Pax3pBSIIKS+/HindIII for 0.8-kb probe; Ptx1pcDNA3/EcoRI for 0.95-kb probe. For immunohistochemistry, HB9 antibody was diluted to 1:8,000. HRP anti-rabbit (The Jackson Laboratory) secondary antibody was used at 1:400, and peroxidase reactions were done according to the manufacturer’s protocol. Other antibodies included mouse anti-hemagglutinin (HA) epitope antibody (HA.11), 1:1,000; rabbit anti-NLI antibody (4508), 1:500; HiFITC anti-mouse (Antibodies, Inc.), 1:300; and Cy3 anti-rabbit (The Jackson Laboratory), 1:300.
RESULTS
Identification of a Novel LIM-Only Gene, LMO4.
To isolate novel LIM domain-containing transcription factors, mouse embryonic day 12 (E12) and 16 (E16) lambda expression libraries were screened with the LID of NLI. Of 2.6 × 106 E12 phage clones screened, two Isl1, two LH-2a (LHX2), one LHX5, and five LMO2 cDNAs were isolated. In addition, 13 phage clones contained cDNAs encoding an as yet uncharacterized LMO protein, which we designated LMO4. The eight positive clones out of 1.3 × 106 E16 phages screened included two LMO1 cDNAs and six LMO4 cDNAs.
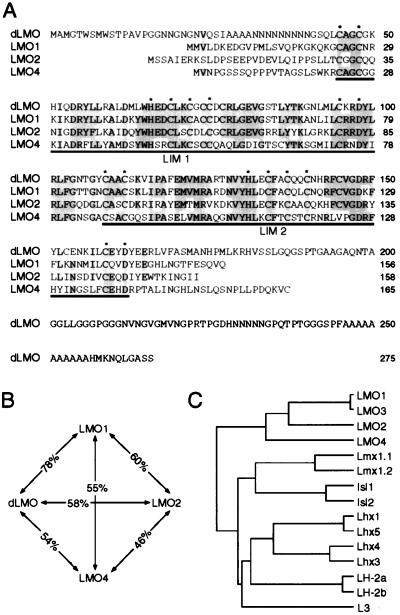
Conceptual translation of the LMO4 cDNA indicated a single ORF encoding a 165-aa protein of approximately 19 kDa, similar in size to the known mammalian LMO proteins, LMO1, LMO2, and LMO3 (18). The human homolog of LMO4 has been deposited in GenBank (accession no. U24576). Sequence comparison of the LMO4 protein to known mammalian and Drosophila LMO proteins indicated that LMO4 is the most distantly related of the LMO family members, with only about 50% amino acid identity within the LIM domains to other LMO proteins (Fig. 1 A and B). In contrast, the LIM domains of LMO1 and the nearly identical LMO3 show 78% identity to the only known Drosophila LMO protein, dLMO (19), and likely represent the vertebrate orthologs of dLMO.
Figure 1.
Sequence analysis of LMO4. (A) Sequence alignment of mouse LMO4 with LMO1 and LMO2, and Drosophila LMO. Shading shows conservation of amino acids, and asterisks indicate the zinc-coordinating residues of each LIM domain. (B) Diagram illustrating relative amino acid sequence identity between the LIM domains of LMO family members. (C) Dendrogram showing genealogical relationships between the LIM domains of LMO and LHX proteins. Comparisons were made using only the LIM domain sequences and inter-LIM region of the rodent representative of each gene, except for LH-2a and LH-2b, which were from chick.
Because LMO proteins display significant sequence homology and similar functional characteristics to the LHX proteins, e.g., nuclear localization, high-affinity interaction with NLI, and assembly into transcription complexes, it is likely that these two subfamilies arose from a gene-duplication event at some point in evolution. Sequence comparison of LIM domains indicates that the LMO proteins are more closely related to the LHX proteins L3 and the LH-2 subgroup (which includes vertebrate LH-2a, LH-2b, the Drosophila protein apterous, and the Caenorhabditis elegans ttx-3) than to any other LHX protein, suggesting that the LMO genes arose from an ancestral LH-2 or L3-like gene (Fig. 1C).
In Vitro and in Vivo Interaction of LMO4 with NLI.
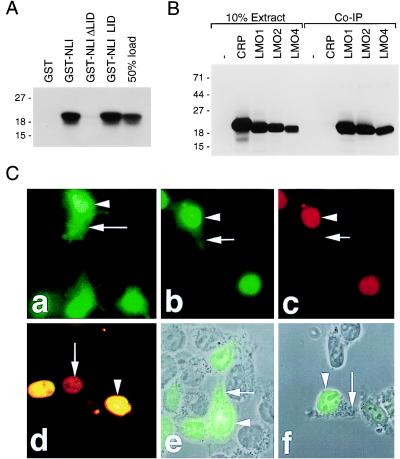
LMO4:NLI interactions were investigated in vitro and in vivo. Glutathione S-transferase (GST) fusion assays in which GST-NLI fusion proteins were incubated with [35S]methionine-labeled LMO4 indicated that LMO4 and NLI associate with high affinity (Fig. 2A), and that the LID of NLI was required and sufficient for interaction with LMO4. When coexpressed with exogenous NLI in embryonic kidney 293 cells, LMO1, LMO2, and LMO4, but not the cytoskeletal-associated LIM protein CRP, coprecipitated efficiently with anti-NLI antibodies (Fig. 2B).
Figure 2.
Association of LMO proteins with NLI in vitro and in vivo. (A) GST-NLI fusions were incubated with in vitro-translated, [35S]methionine-labeled LMO4, and 50% of the in vitro-translated material used in the binding reaction was loaded separately for estimation of binding affinity. Interaction was visualized by SDS/PAGE and fluorography. (B) In vivo association of LMO proteins with NLI. Anti-NLI immunoprecipitation and anti-HA epitope immunoblotting of whole-cell extracts from cotransfected 293 cells. Ten percent of the extracts was loaded separately to show relative expression. (C) Nuclear retention of LMO proteins depends on NLI. Immunohistochemistry using an anti-HA epitope antibody (Babco, Richmond, CA) showed diffuse localization of HA-tagged LMO2 (a) and LMO4 (e) in transiently transfected 293 cells. Cotransfection of untagged NLI with LMOs resulted in nuclear retention of LMO2 (b) and LMO4 (d and f). NLI was localized to the nucleus (c and d). (d) Merging of NLI and HA-LMO4 staining shows codistribution in cell nuclei (arrowhead) and endogenous expression of NLI (arrow). For e and f, transmitted light images of differential interference contrast optics were captured in registration with the fluorescent signals.
The small size of LMO proteins suggests that they should be capable of freely traveling in and out of the nucleus; however, the lack of a nuclear localization signal (NLS) sequence in the LIM-only transcription factors suggests that they lack an intrinsic mechanism for nuclear retention. It is therefore unclear how LMO proteins are localized predominately in the nucleus (7). Using an anti-HA epitope antibody, immunocytochemistry of 293 cells transfected with HA-tagged LMO2 or LMO4 showed distribution in both the nucleus and cytoplasm (Fig. 2C, a and e). However, upon cotransfection of the LMO cDNAs with NLI, anti-HA and anti-NLI immunocytochemistry revealed a predominantly nuclear localization (Fig. 2C, b–d, f). NLI contains at least two NLS sequences and is a nuclear protein (10). Since cotransfection of NLI with LMO cDNAs promoted nuclear retention of LMO proteins, the partial nuclear distribution of LMO2 and LMO4 in cells not transfected with NLI is likely a result of endogenous NLI (Fig. 2C, d). These results indicate that one function of NLI in cells is to maintain the nuclear localization of LMO proteins.
Differential LMO mRNA Expression in Adult Mouse Tissues.
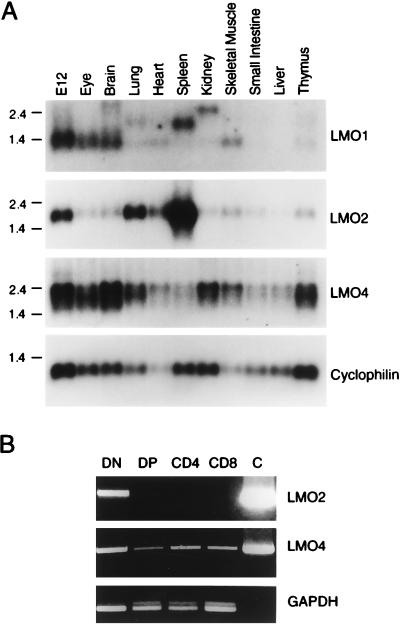
To compare the tissue distribution of LMO4 gene expression with other LMO genes, Northern blots of poly(A)+ RNA isolated from E12 mouse embryos and various adult mouse tissues were compared. While low levels of LMO1 expression could be detected only in the E12 embryo, eye, brain, and skeletal muscle, LMO2 expression was highest in the E12 embryo, spleen, and lung as noted previously (5, 18, 20) (Fig. 3A). The signals in the lung and spleen lanes of the LMO1 blot are the result of residual hybridization of the LMO2 probe. Low-level expression of LMO2 was detectable in the thymus as noted previously (20). In contrast to the relatively restricted pattern of expression of LMO1 and LMO2, LMO4 mRNA was more widely expressed, with highest levels in the eye, brain, kidney, and, intriguingly, the thymus. The mRNA expression of LMO genes in adult tissues is overlapping, but clearly distinct.
Figure 3.
(A) Northern analysis of various adult mouse tissues and the E12 embryo shows combinatorial and complementary patterns of expression of LMO genes. (B) Reverse transcription–PCR analysis of thymic sublineages. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) estimates relative quantities of cDNAs in each lane. C is a control PCR product from 0.1 μg of the appropriate template cDNA. DN, double-negative (CD3−CD4−CD8−); DP, double-positive (CD4+CD8+).
Because unregulated expression of LMO1 (21) and LMO2 (6) in T cells results in leukemogenesis, we examined the expression of LMO4 in adult thymus tissue at a cellular level. In situ hybridization showed widespread expression of LMO4 throughout the thymus, consistent with expression in the lymphoid lineage (not shown). To identify the types of lymphoid cells that express LMO4 and LMO2, we isolated the four major thymic subsets: immature blast cells (DN), negative for CD3, CD4, and CD8; CD4+CD8+ double-positive cells (DP); and single-positive (SP) mature CD4+ or CD8+ cells. The DN cells are predominantly immature T cells, but may contain trace amounts of non-T cells of the lymphoid lineage. As shown in Fig. 3B, LMO4 was expressed in all of the four major thymic subsets, while LMO2 expression was restricted to the immature blast cells. Therefore, LMO4 and LMO2 are coexpressed in proliferating blast cells, but differentially regulated in double-positive and single-positive subsets.
LMO4 Expression During Somitogenesis.
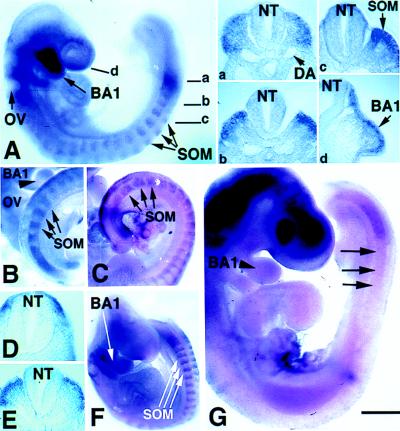
To analyze the temporal and spatial patterns of LMO4 expression in the embryo, an LMO4 probe was used in in situ hybridization experiments in whole mount and on sections. In the 16 somite-stage embryo (E9.0), LMO4 expression was distributed rostrally in migratory cranial crest within the branchial arches, and caudally in dorsal paraxial mesoderm (Fig. 4A). In paraxial mesoderm, expression was initially restricted dorsally (Fig. 4A, a). The metameric pattern of expression seen in the presomitic mesoderm was maintained in the somite, as LMO4 was restricted to the rostral portion of each somite (Fig. 4A, b and c). LMO4 expression persisted in paraxial mesoderm at E9.5, with highest levels observed in the rostral portion of the first few newly formed somites (Fig. 4B). At anterior axial levels, LMO4 expression was restricted to cells adjacent to the neural tube (Fig. 4D). Thus, LMO4 is activated in cranial neural crest cells and dorsal paraxial mesoderm.
Figure 4.
Distribution of LMO4 mRNA in cranial crest and paraxial mesoderm revealed by whole-mount in situ hybridization. (A) Sixteen somite-stage embryo exhibits LMO4 expression (blue or brown stain) in cranial neural crest migrating into the branchial arches and in rostral somite. Sections at indicated axial levels show expression in dorsal paraxial mesoderm (a), prospective dermomyotome (b), dermomyotome (c), and migratory cranial neural crest in the mandibular component of the first branchial arch (BA1) (d). Otic vesicle (OV), neural tube (NT), and somites (SOM) are indicated by arrows. DA, dorsal aorta. (B) At E10, LMO4 expression in paraxial mesoderm and in rostral somite halves persists at a caudal axial level (E) and is restricted medially at anterior levels to cells adjacent to the neural tube (D). (B) As indicated by arrowhead, LMO4 expression was restricted to ventral mandibular arch by this stage. (C) Pax3 expression is not restricted, but is distributed throughout the dermomyotome. (F) LMO4 activation in Splotch mice is similar to normal embryos in migratory cranial neural crest cells and unsegmented paraxial mesoderm, yet somitic expression is not restricted medially. (G) In Mesp2 mutant embryos, activation of LMO4 in unsegmented mesoderm appears normal. However, expression is lost in rostral somite (arrow). Expression in mandibular arch is detected at a diminished level. Staining in the head region is an artifact caused by probe trapping. [Bar = 200 μm (A, B, C, and G) and 400 μm (D and F).]
We compared the pattern of expression of LMO4 with that of Pax3, a paired-type Hox gene known to be expressed in neural crest and the dermomyotome (22, 23). Unlike LMO4, which is restricted to the rostral portion of newly formed somites, and restricted medially in mature somites (Fig. 4B), Pax3 expression persisted throughout the dermomyotome in mature somites (Fig. 4C). The observation that LMO4 and Pax3 are coexpressed initially raised the possibility that LMO4 might be regulated by Pax3. In Splotch mutants, which lack functional Pax3, LMO4 was activated normally in unsegmented dorsal mesoderm and in migratory cranial crest (Fig. 4F). However, the domain of LMO4 expression in mature somites persisted laterally in Splotch embryos (Fig. 4F), raising the possibility that Pax3 may normally restrict expression of LMO4.
LMO4 activation in Mesp2 mutants was examined to discern whether LMO4 expression is linked to somite segment polarity or differentiation of somite lineages. Mesp2 is a basic helix–loop–helix (bHLH) transcription factor required for normal rostral–caudal segmental polarity of somite tissue but not differentiation of somitic lineages, such as muscle (24). LMO4 was expressed in presomitic mesoderm of Mesp2 mutant embryos. However, activation of LMO4 in the rostral portion of newly formed somites was not detectable (Fig. 4G). Therefore, maintenance of LMO4 expression in somite requires Mesp2 activity. Since Mesp2 mutants have somitic derivatives such as muscle and sclerotome, we infer that LMO4 is not required for differentiation of somitic lineages.
Expression of LMO Genes in Limb Bud and Mandibular Arch.
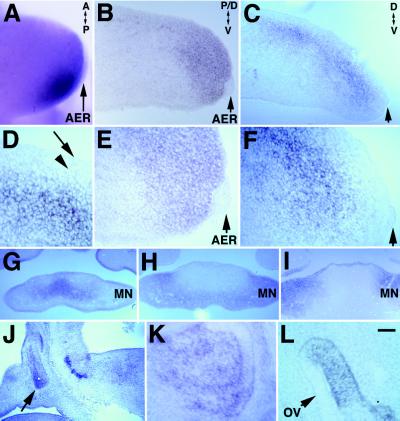
During limb outgrowth and patterning, both LMO2 and LMO4 have expression domains that overlap with progress zone mesenchyme, with LMO2 being expressed in distal and posterior mesenchyme (Fig. 5A and B), and LMO4 in dorsal mesenchyme. At E11.5, LMO4 expression was detected in dorsal mesenchyme extending along the proximal–distal axis of the limb bud (Fig. 5C). While LMO4 transcripts were not detected in mesenchyme subjacent to overlying ectoderm (Fig. 5 D and F), LMO2 transcripts were detected in mesenchyme subjacent to ectoderm and to the apical ectodermal ridge (AER) (Fig. 5E).
Figure 5.
Expression of LMO4 and LMO2 in limb tissue and expression of LMO4 in ventral mandible, anterior pituitary, and otic vesicle revealed by nonisotopic in situ hybridization to tissue sections. (A, B, and E) LMO2 is activated in posterior–distal mesenchyme subjacent to the AER (arrow) at E9.5 (A) and persists at E11.5 (B and E). E is a ×2.5 magnification of tissue in B. (C, D, and F) Activation of LMO4 occurs in dorsal mesenchyme in E11.5 limb (C) that does not contact ectoderm (arrow) (D and F). Arrowhead marks subjacent mesenchyme. D and F are a ×2.5 magnification of C. Arrow in C and F indicate AER. (G) Near-adjacent sections show that activation of LMO4 in E11.5 ventral mandible is restricted medially, while Pax3 (H) and Ptx1 (I) expression overlap in a complementary domain laterally. (J and K) Sagittal section through the head shows LMO4 expression in anterior pituitary tissue at E11.5 (J) and E14.5 (K). Expression of LMO4 persists in distinct regions of the otic vesicle (OV) at E11.5. [Bar = 60 μm (A), 50 μm (B, C, G–J), and 20 μm (D–F, K, L).]
In the developing jaw, LMO4 was expressed at high levels in both ventral and dorsal mandibular mesenchyme at E8.5 (Fig. 4A, d), yet between E9.5 and E11.5, expression was progressively restricted ventrally to the transitory mesenchyme that ultimately joins the left and right components of mandibular arch (Figs. 4B and 5G; data not shown). Pax3 and Ptx1 were coexpressed in a domain associated with differentiation of cartilage and muscle (25) that was complementary to the LMO4 expression domain (Fig. 5 G–I). Therefore, LMO4 expression was excluded from differentiating tissue and defined the transitory mesenchyme that joins the left and right mandibular arches at the midline.
LMO4 expression also was detected in nasalpharyngeal ectoderm and maxillary mesenchyme during mergence of the frontonasal process (Fig. 5G; data not shown). LMO4 is activated in other tissues, including early motor neurons of the ocularmotor nerve, hindbrain motor neurons, glial cells associated with the optic nerve and cranial nerves, anterior pituitary, otic vesicle, and later in forebrain neurons (Fig. 5 J–L; data not shown). LMO4 activation in the anterior pituitary first was detected at E11.5 and persisted until E14.5, the last time examined (Fig. 5 J and K). During ear development, expression of LMO4 first appeared in the lateral wall of the closing otic vesicle and persisted in the semicircular canal primoridia at E11.5 (Fig. 5L).
LMO4 Expression in Motor Neurons and Schwann Cell Progenitors.
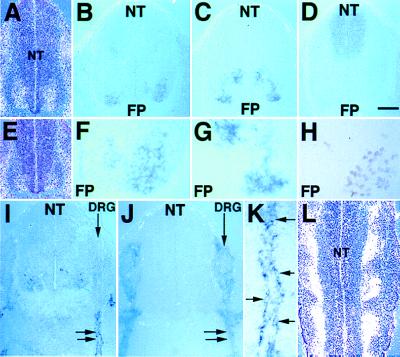
At E11.5, the domain of LMO4 expression extended more dorsally in rostral spinal cord relative to caudal levels (Fig. 6 A and E). Comparison of adjacent sections in the lumbar spinal cord shows that LMO4 expression (Fig. 6 B and F) overlapped with LHX3 expression in the medial subdivision of the median motor column (MMCm), but not with LHX3 expression in interneurons (Fig. 6 C and G). Expression of LMO4 in cells slightly more dorsal to the MMCm overlaps with the pan-motor neuronal marker HB9 (Fig. 6H). Expression of LMO4 in apically located paraventricular cells (Fig. 6B) does not overlap with the domain of Pax3 expression (Fig. 6D). LMO4 expression in motor neurons persisted at later times (data not shown). These data show that LMO4 is activated in spinal cord motor neurons soon after neuroblast migration, during the period of neurite outgrowth.
Figure 6.
Activation of LMO4 in motor neurons and Schwann cell progenitors. (A and E) In situ hybridization of the 35S-labeled LMO4 probe to transverse sections counterstained with hematoxylin shows that the domain of LMO4 neural tube (NT) expression (white grains) is broad at a rostral axial level (A) relative to caudal axial level (E). In situ hybridization of digoxygenin-labeled probes to adjacent transverse sections through the E11.5 lumbar spinal cord shows overlap of the LMO4 expression pattern (Blue) (B and F) with motor neuron expression of LHX3 (C and G), but not with LHX3-expressing interneurons. HRP immunohistochemistry shows that the expression domain of the pan-motor neuronal marker HB9 (H) on adjacent sections partially overlaps with LMO4 and LHX3 expression domains. FP, floor plate. F and G are ×2.5 magnifications of B and C, respectively. Apical paraventricular cells that express LMO4 (B) do not overlap with the Pax-3 expression domain (D). (I) At E11.5, activation of LMO4 occurs in cells tightly associated with spinal nerves, indicated by double arrows. (J) Sox10 expression in Schwann cells overlaps with LMO4 expression (I). K is a ×3 magnification of the tissue in I. Arrows indicate individual cells decorating the spinal nerve that express LMO4. (L) Longitudinal section through the E11.5 neural tube hybridized with 35S-labeled LMO4 probe and counterstained with hematoxylin shows expression (white grains) at the exit points for spinal nerves. [Bar = 50 μm (A–E, J), 20 μm (F– H, L), 60 μm (I), and 12 μm (K).]
LMO4 transcripts also were present in individual cells tightly associated with cranial and spinal nerves (Fig. 6 I, K, and L; data not shown). The glia marker Sox10 (26) and LMO4 have identical expression patterns along nerve fibers, but differ within the dorsal root ganglia (DRG), where Sox10, but not LMO4, is present at this stage (Fig. 6 I and J). Overlapping expression of LMO4 and Sox10 indicates that LMO4 is activated in Schwann cell progenitors after their emergence from the DRG.
DISCUSSION
NLI:LIM Domain Complexes.
LMO and LHX proteins form tetrameric complexes with NLI that are proposed to regulate gene expression (1). Because combinatorial association of LHX proteins is mediated by dimeric NLI (13), the widespread abundance of NLI provides for tetrameric regulatory complexes between LHX and LMO proteins. We show several cases in which LMO and LHX gene expression overlap. Although it is difficult to make a meaningful assessment of the biological relevance for NLI:LMO:LHX complexes, expression of LMO4 and LMO2 during early embryogenesis suggests a possible role in maintenance of an undifferentiated state, potentially by disruption of LHX activity.
LMO4 expression in dorsal limb mesenchyme partially overlaps with the LHX gene Lmx1, which is thought to dorsalize the limb in response to Wnt7a (for review, see ref. 27). The time and location of LMO4 activation implicate LMO4 as a possible gene target of Lmx1. NLI is widely expressed in E10.5 limb mesenchyme and, like LMO4 and LMO2, is restricted to perichondral tissue during digit formation at E14.5 (data not shown). Since synergistic interactions between Lmx1 and E47 are disrupted by NLI (28), formation of complexes containing NLI and Lmx1 in subectodermal limb mesenchyme are likely to modulate Lmx1 activity during limb dorsalization. In deeper mesenchyme, LMO4 likely would modify the NLI:Lmx1 complex. Identification of LMO4 as both a potential gene target and transcriptional modulator of Lmx1 activity should help define the role of Lmx1 during limb patterning.
In the spinal cord, motor neurons are parsed into an array of functionally distinct motor columns, with each column defined by combinatorial expression patterns of LHX genes (29). We show that the spinal cord is further imbricated by expression of LMO4. The activation of LMO4 in E10.5 motor neurons occurs after the onset of high levels of Isl1 and NLI expression at E9.5 (10). Therefore, LMO4 likely modulates the transcriptional activity of NLI:Isl1 complexes after the initial role of Isl1 in the generation of motor neurons.
LMO Expression in Uncommitted Tissue.
The activation of LMO2 adjacent to the zone of polarizing activity (ZPA) and subjacent to the AER defines a unique population of mesenchyme that connects the AER and ZPA signaling centers of the limb. The lack of commitment within the progress zone that underlies the AER is linked to continued outgrowth of the limb, and both qualities are stimulated by reciprocal interactions of the ZPA with the AER (27). A potential role for LMO2 in blocking commitment of progress zone-associated tissue to regional fates conforms with the functional precedent for LMO2 in inhibition of cellular differentiation (30, 31).
Schwann cell progenitors activate LMO4 after making axonal contact, during the period in which Schwann cell differentiation is delayed (32). Schwann cell progenitors within the DRG remain multipotent, capable of forming both Schwann and pigment cells (33). Expression of LMO4 first is detected after Schwann cell progenitors emerge from DRG and contact spinal nerves, raising the possibility that axonal contact activates LMO4.
LMO4 and LMO2 are regulated differentially during the development of thymic subsets. A role for LMO2 in promotion of blast proliferation and inhibition of T lymphocyte differentiation was proposed based on analysis of transgenic animals (31). Our data showing blast cell expression of LMO2 suggest a role for LMO2 in normal T cell proliferation. The demonstration that ectopic expression of either LMO1 or LMO2 leads to tumorigenesis in T cells (6, 21) suggests that these structurally related proteins perform a similar function in cell growth. The normal expression of LMO4 in all thymic subsets and coexpression of LMO2 and LMO4 in proliferating blast cells suggest that T cell proliferation or differentiation may be determined by the level of LMO expression.
LMO4 activation by migratory cranial neural crest cells and transitory mesenchyme defines a stage when these tissues are uncommitted to regional fates. Cranial neural crest cells are generated at the boundary between ectoderm and neural epithelia (34), yet are prevented from becoming either ectoderm or neural epithelial derivatives. Activation of LMO4 expression at the onset of migration marks the time of divergence between the cranial neural crest cell lineage and the ectodermal and neural tube cell lineages. LMO4 expression similarly distinguishes transitory mesenchyme, which establishes continuity between the core mesenchymal components of each mandibular process, from neighboring differentiating mesenchyme. Therefore, LMO4 expression in the mandibular arch characterizes uncommitted tissue that supports morphological transformations essential for facial development.
Regulation of LMO4.
Lack of LMO4 expression in Mesp2 mutant embryos demonstrates that LMO4 activation is regulated during the establishment of somite formation and that somites in Mesp2 mutants lack a rostral character. Although the significance of rostral–caudal differences in the dermomyotome is unclear, the rostral–caudal differences in newly formed somites are likely to mediate the formation of somite boundaries. Maintenance of both LMO4- and Notch-signaling molecules during somite development requires Mesp2, but it remains to be determined whether Notch signaling and LMO4 act together or in parallel. Alternatively, LMO4 activation could be downstream of FGFR1 activation, since FGFR1 somitic expression also is absent in Mesp2 mutant mice (24).
The diversity seen in expression of LMO4 may be indicative of a fundamental mechanism of gene regulation that is common to separate patterning events. In addition to LMO4, signaling mechanisms that involve BMPs, fibroblast growth factors, sonic hedgehog, and wingless proteins are reiterated during patterning of the face, limb, and somite (27, 36–38). Therefore, via interaction with NLI, LMO4 may modulate the activity of transcriptional complexes in response to highly conserved signaling mechanisms that pattern the early embryo.
Acknowledgments
We are grateful to the following people for helpful discussions and assistance: David Schwarz for assistance with isolation of thymic sublineages, Sam Pfaff for the HB9 antibody, and Martyn Goulding for the Splotch mice and Pax3 cDNA. We thank Heiner Westphal for the Lhx3 cDNA, Michael Wegner for the Sox10 cDNA, and Jacque Drouin and Pamela Mellon for the Ptx1 cDNA. These studies were supported by National Institutes of Health Grant DK13149. D.A.K. is supported by National Institutes of Health Training Grant T32HL07770 and L.W.J. is supported by National Institutes of Health Training Grant DK07541.
ABBREVIATIONS
- LHX
LIM homeobox
- LMO
LIM-only family of nuclear LIM proteins
- NLI
nuclear LIM interactor (also named Ldb1, LIM domain-binding protein
- CLIM
cofactor of LIM homeodomain proteins)
- LID
LIM interaction domain of NLI
- AER
apical ectodermal ridge
- ZPA
zone of polarizing activity
- DRG
dorsal root ganglia
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF074600 (mouse LMO4)].
References
- 1.Dawid I B, Breen J J, Toyama R. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 2.Shawlot W, Behringer R R. Nature (London) 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- 3.Pfaff S L, Mendelsohn M, Stewart C L, Edlund T, Jessell T M. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 4.Boehm T, Foroni L, Kaneko Y, Perutz M F, Rabbitts T H. Proc Natl Acad Sci USA. 1991;88:4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Royer-Pokora B, Loos U, Ludwig W-D. Oncogene. 1991;6:1887–1893. [PubMed] [Google Scholar]
- 6.Fisch P, Boehm T, Lavenir I, Larson T, Arno J, Forster A, Rabbitts T H. Oncogene. 1992;7:2389–2397. [PubMed] [Google Scholar]
- 7.Warren A J, Colledge W H, Carlton M B, Evans M J, Smith A J, Rabbits T H. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 8.Yamada Y, Warren A J, Dobson C, Forster A, Pannell R, Rabbitts T H. Proc Natl Acad Sci USA. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wadman I A, Osada H, Grutz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jurata L W, Kenny D A, Gill G N. Proc Natl Acad Sci USA. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agulnick A D, Taira M, Breen J J, Tanaka T, Dawid I B, Westphal H. Nature (London) 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- 12.Bach I, Carriere C, Ostendorff H P, Andersen B, Rosenfeld M G. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 13.Jurata L W, Pfaff S L, Gill G N. J Biol Chem. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- 14.Morcillo P, Rosen C, Baylies M K, Dorsett D. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaeren-Wiemers N, Gerfin-Moser A. Histology. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 16.Angerer L M, Toler M H, Angerer R C. In: In Situ Hybridization–Applications to Neurobiology. Valentino K L, Eberwine J H, Barchas J D, editors. Oxford: Oxford Univ. Press; 1987. pp. 42–70. [Google Scholar]
- 17.Wilkinson D G, Nieto M A. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 18.Foroni L, Boehm T, White L, Forster A, Sherrington P, Liao X B, Brannan C I, Jenkins N A, Copeland N G, Rabbitts T H. J Mol Biol. 1992;226:747–761. doi: 10.1016/0022-2836(92)90630-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhu T H, Bodem J, Keppel E, Paro R, Royer-Pokora B. Oncogene. 1995;11:1283–1290. [PubMed] [Google Scholar]
- 20.Neale G A M, Mao S, Parham D M, Murti K G, Goorha R M. Cell Growth Differ. 1995;6:587–596. [PubMed] [Google Scholar]
- 21.McGuire E A, Rintoul C E, Sclar G M, Korsmeyer S J. Mol Cell Biol. 1992;12:4186–4196. doi: 10.1128/mcb.12.9.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goulding M, Sterrer S, Fleming J, Balling R, Nadeau J, Moore K J, Brown S D J, Steel K P, Gruss P. Genomics. 1993;17:355–363. doi: 10.1006/geno.1993.1332. [DOI] [PubMed] [Google Scholar]
- 23.Goulding M, Lumsden A, Paquette A J. Development. 1994;120:957–971. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- 24.Saga Y, Hata N, Koseki H, Taketo M M. Genes Dev. 1997;11:1827–1839. doi: 10.1101/gad.11.14.1827. [DOI] [PubMed] [Google Scholar]
- 25.Lanctot C, Lamolet B, Drouin J. Development. 1997;124:2807–2817. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- 26.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wenger M. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson R L, Tabin C J. Cell. 1997;90:979–990. doi: 10.1016/s0092-8674(00)80364-5. [DOI] [PubMed] [Google Scholar]
- 28.Jurata L W, Gill G. Mol Cell Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuchida T, Ensini M, Morton S B, Baldassare M, Edlund T, Jessell T M, Pfaff S L. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 30.Visvader J E, Mao X, Fujiwara Y, Kyungmin H, Orkin S H. Proc Natl Acad Sci USA. 1997;94:13707–13712. doi: 10.1073/pnas.94.25.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neale G A M, Rehg J E, Goorha R M. Blood. 1995;86:3060–3071. [PubMed] [Google Scholar]
- 32.Monuki E S, Weinmaster G, Kuhn R, Lemke G. Neuron. 1989;3:783–793. doi: 10.1016/0896-6273(89)90247-x. [DOI] [PubMed] [Google Scholar]
- 33.Stocker K M, Sherman L, Rees S, Ciment G. Development. 1991;111:635–645. doi: 10.1242/dev.111.2.635. [DOI] [PubMed] [Google Scholar]
- 34.Selleck M A, Bronner-Fraser M. Development. 1995;121:525–538. doi: 10.1242/dev.121.2.525. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen M M, Abate-Shen C. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hogan B L M. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 37.Wall N A, Hogan B L M. Mech Dev. 1995;53:383–392. doi: 10.1016/0925-4773(95)00453-x. [DOI] [PubMed] [Google Scholar]
- 38.Richman J M, Tickle C. Dev Biol. 1992;154:299–308. doi: 10.1016/0012-1606(92)90069-s. [DOI] [PubMed] [Google Scholar]