Abstract
Phylogenies based on the inheritance of shared derived characters will be ambiguous when the shared characters are not the result of common ancestry. Such characters are called homoplasies. Phylogenetic analysis also can be problematic if the characters have not changed sufficiently, as might be the case for rapid or recent speciations. The latter are of particular interest because evolutionary processes may be more accessible the more recent the speciation. The repeated DNA subfamilies generated by the mammalian L1 (LINE-1) retrotransposon are apparently homoplasy-free phylogenetic characters. L1 retrotransposons are transmitted only by inheritance and rapidly generate novel variants that produce distinct subfamilies of mostly defective copies, which then “age” as they diverge. Here we show that the L1 character can both resolve and date recent speciation events within the large group of very closely related rats known as Rattus sensu stricto. This lineage arose 5–6 million years ago (Mya) and subsequently underwent two episodes of speciation: an intense one, ≈2.7 Mya, produced at least five lineages in <0.3 My; a second began ≈1.2 Mya and may still be continuing.
Cladistics is a phylogenetic approach for classifying organisms into taxa based on shared inherited characters (1). The emphasis on inherited couples taxonomic classification to the evolutionary history of the examined taxa. This makes cladistics intellectually appealing since phylogeny is based on genealogy. The shared characters can range from classical morphological and biochemical to molecular sequence data.
However, the major problem for cladistics is determining whether a shared character is inherited or arose independently because of convergence, parallelisms, or reversion to an ancestral state. Noninherited shared characters are called homoplasies, and they can lead to multiple, equally likely phylogenetic trees or, in extreme cases, a single incorrect tree (e.g., see ref. 2). An additional problem occurs for rapid speciations because phylogenetic characters may not have changed sufficiently (3). Recently we (4–6) and others (7–11) have shown, respectively, that L1 (LINE, long interspersed) and SINE (short interspersed) repeated DNA elements apparently are homoplasy-free characters. However, in most cases the repeated elements have been used differently as phylogenetic characters. Although the phylogenetic distribution of distinct SINE families has been informative (11), usually the presence or absence of SINE element insertions at particular loci has been used as a phylogenetic character. While L1 elements also can be used this way, the presence or absence of distinct multicopy L1 subfamilies has been scored as the phylogenetic character.
This difference stems from the distinct biological properties of these elements. L1 elements are prolific, self-replicating mammalian retrotransposons that rapidly generate distinct novel subfamilies consisting mostly of defective (pseudo) copies (see legend to Fig. 1). The defective subfamily members are retained in the genome and diverge from each other with time at the pseudogene (neutral) rate. The rapid generation of novel L1 characters keeps pace with speciation, and the sequence divergence of the various defective subfamily members theoretically permits the dating of the speciations (4). By contrast, although SINE elements can be organized into subfamilies, they are not self-replicating and there are not enough distinct SINE families to generate high-resolution trees (11). Although individual SINE insertions are very robust phylogenetic characters and can generate detailed phylogenies, they cannot be used to date phylogenetic events (12).
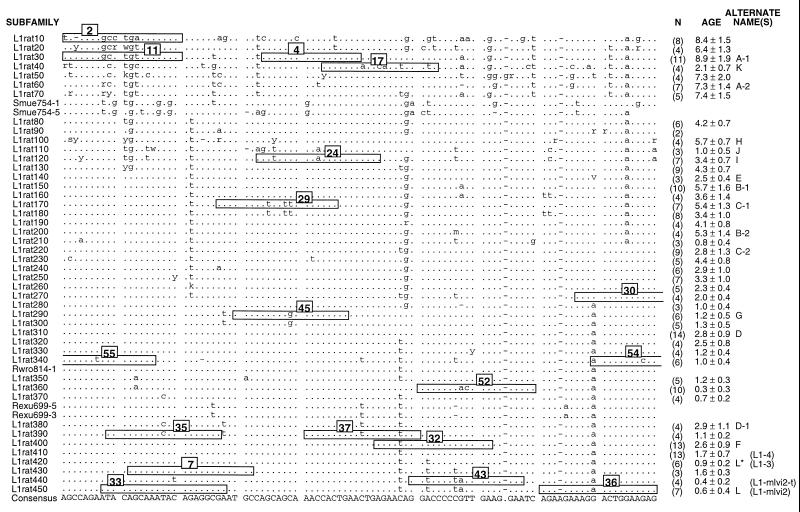
Figure 1.
Rat L1 subfamilies. This is an alignment of the consensus sequences calculated for 45 rat L1 subfamilies and the individual sequence of 5 additional L1 elements. Only 100 bp of the 215- to 320-bp region of the 3′ UTR that was sequenced is shown here. All mammalian L1 elements contain four regions: a 5′ UTR involved in regulation; ORF I, which encodes an RNA-binding protein; ORF II, which encodes a reverse transcriptase; and the 3′ UTR. As explained elsewhere, the evolution of the 3′ UTR appears to occur rapidly enough to make it a useful source of phylogenetic characters for analyzing recent or rapid speciations (4). The names of the subfamilies (or individual elements) are given on the left, and the number (N) of members of each subfamily and its approximate age (in My) are given on the right. Alternate names also are listed on the right: letters for cross-reference to Fig. 2 and in parentheses are previously used designations (e.g., refs. 6 and 19). The dots indicate identity with the consensus calculated from the listed sequences, the dashes indicate gaps, and the letters indicate differences. The numbered boxes indicate the sequence of the oligonucleotide hybridization probes derived from this part of the alignment. For oligonucleotides 55, 54, and 30, the sequence extends a few bases beyond the displayed alignment. This alignment begins at base 10 of our previously published partial alignment of this region (6).
Here we demonstrate that the L1 phylogenetic character can determine and date phylogenetic events within Rattus sensu stricto. These rodents consist of ≈50 very closely related taxa that evolved very recently and have been largely refractory to phylogenetic analysis (13–16). We found that the Rattus sensu stricto lineage, which we redefine partially here, emerged ≈7.5–5.5 million years ago (Mya). Rattus sensu stricto then underwent two intense speciations: one occurred ≈2.7 Mya and generated five Rattus lineages in less than 0.3 My; a second began ≈1.2 Mya and may still be continuing.
MATERIALS AND METHODS
Biological Specimens.
The rodent samples (except R. norvegicus from New York and Mus musculus domesticus, a laboratory strain), were from the collection of the Institut des Sciences de l’Evolution of Montpellier II (17). The species names, registry numbers, geographical localities, and collectors of the different specimens have been described (6). We follow the nomenclature and taxonomy presented in ref. 18 with the following exceptions as explained in ref. 6: Niviventer niviventer, Rattus flavipectus, R. cf moluccarius, and R. satarae. Of the 26 species of Rattus sensu lato examined, 4 belong to the Maxomys genus, 4 belong to Niviventer, 2 belong to Leopoldamys, 1 each belong to Berylmys, Sundamys, and Bandicota, and 13 belong to Rattus. For outgroup comparisons, we examined four Murinae species: Mus musculus domesticus, Aethomys namaquensis, Thamnomys gazellae, and Conilurus penicillatus; and four non-Murinae species: Cricetomys gambianus, Tatera indica, Akodon torques, and Arvicola terrestris.
General Techniques.
DNA was purified from preserved tissues of the above specimens as described (6). The DNA was digested with Sau3AI and NlaIII, whose sites are highly conserved in the 3′ untranslated region (UTR) of rat L1 elements (see legend to Fig. 1 and refs. 6 and 19) and define a 215-bp fragment that was purified by gel electrophoresis and ligated to the dephosphorylated BamHI site of pUC19 as described (6). Transfected bacteria were screened for L1-containing clones by hybridization with a fragment of the 3′ UTR at moderate stringency (6). DNA sequencing, blotting of restriction endonuclease-digested genomic DNA, and blot hybridizations with oligonucleotide probes were carried out by using standard procedures described in ref. 6 or refs. 20–22. The 206 sequences that had not been reported previously have been deposited in GenBank/EMBL (accession nos. AJ004354–AJ004559). Generally, these sequences correspond to the expected ≈215 bp of the 3′ UTR. However, occasionally both longer fragments (≈320 bp) and shorter fragments (<≈150 bp) were sequenced.
DNA Sequence Analysis.
Sequences were manipulated with either the must (23) or GCG programs (Version 8, Genetics Computer Group, Madison, WI). The sequences were aligned and roughly sorted into groups of related sequences by using the neighbor-joining method (24). Consensus sequences were calculated for every group of three or more sequences and compared with the members of the group to determine whether additional subsets of distinct sequences were present. Any such subsets were separated into new groups, their consensus sequences were calculated, and the above comparison was repeated. After several iterations of this process we reduced all but 5 of the 245 sequences used for this study into 45 L1 subfamilies, which are listed in Fig. 1. Comparison between the consensus sequence of each subfamily with the overall consensus sequence derived from them revealed nucleotide differences that distinguished each subfamily (Fig. 1). Oligonucleotides cognate to these nucleotide differences were hybridized to genomic DNA in the presence of one or more competitor oligonucleotides which were cognate to either the overall consensus sequence or to that of closely related subfamilies (6). We also defined some oligonucleotide probes on L1 elements not grouped into subfamilies or to regions of L1 subfamily members that differed from the subfamily consensus sequence (i.e., private changes). The sequences of all of the oligonucleotides and their competitors (when defined), the entire alignment of the consensus sequences of all of the subfamilies, and the L1 elements assigned to each subfamily are available either from the authors or by anonymous file transfer protocol (ftp) from helix.nih.gov in the file Verneau.doc. Maximum parsimony was carried out by using the paup 3.1.1 program (25).
Calculation of Subfamily Age.
Subfamily age was determined from the sequence divergence (genetic distance) between every pair of subfamily members (excluding those <150 bp) after correcting for superimposed mutations using the 2-ρ method of Kimura (26). The average pairwise distance for each subfamily was converted into years by using the pseudogene (neutral) base substitution rate of 1.1% per million years (My) calculated for rodent genomes (27). We obtained the same value by using the divergence of an ancestral murine L1 family, L1mur-1 (previously called Lx), and 12 My as the time of the murine radiation that was estimated from the fossil record (refs. 28 and 29 and references therein).
RESULTS
Fig. 1 shows part of the alignment of the consensus sequences of 45 rat L1 subfamilies identified here and the oligonucleotide probes defined from this part of the alignment. Subfamilies (e.g., L1rat290 and L1rat300, L1rat380 and L1rat390) that are identical in this region of the alignment are clearly distinguished in regions not displayed in Fig. 1. Oligonucleotide probes were hybridized to blots of genomic DNA that had been digested with various restriction endonucleases (Materials and Methods and refs. 4 and 6). As shown previously and discussed in detail (4, 6), these reactions generally revealed distinctive patterns of hybridized bands that greatly enhanced both the specificity and the information content of the hybridizations.
Since the L1 phylogenetic character is the result of a hybridization reaction, we refer to each L1 character by the name of the oligonucleotide hybridization probe rather than by the name of the L1 subfamily upon which the oligonucleotide was defined. This avoids confusion because an ancestral L1 oligonucleotide character can be retained in present-day mammals in two ways. First, old L1 subfamilies are not cleared from the genome. Therefore, the oligonucleotide characters defined on old L1 subfamilies will be retained until they are no longer detectable by hybridization because of the accumulation of random mutations as the old L1 elements “age” as pseudogenes.
Second, an ancestral character can be retained in the younger, modern L1 subfamilies that evolved from an ancestral L1 family, perhaps as a result of selection (for the functional properties of the retained sequence) or by the recombination of a modern active element with an older element (19). For example, Fig. 1 shows that oligonucleotide 4 is present in the ≈9-My-old L1rat30 subfamily and also has been retained in numerous modern L1 subfamilies (e.g., the ≈0.5-My-old L1rat440 subfamily and many others). Because of this and the fact that we have sampled only a very small fraction of genomic L1 elements, we also would expect that an oligonucleotide character defined only on a young L1 subfamily could have a wider phylogenetic distribution than predicted by the age of the subfamily. For example, this would be have been the case for oligonucleotide 4 if we only had sampled members of the L1rat440 subfamily. Several of the oligonucleotide characters defined only on young subfamilies have this property.
Finally, although some oligonucleotide characters embrace multiple diagnostic nucleotides, others are cognate to only a single-base difference. Since a one-base difference could occur by chance as a private change in any genomic L1 element, this would result in homoplasy due to parallelism if the occurrence was detected as a hybridization signal with the oligonucleotide probe. We avoided this problem by using an amount of genomic DNA (≈100 ng) sufficient only to generate a hybridization signal when ≥20 copies of an L1 sequence were present and ignored signals corresponding to less than ≈100 copies (4, 6). Parallelism due to the independent amplification of L1 subfamilies that coincidentally hybridize to the same oligonucleotide probe is discussed below.
Fig. 2 shows the phylogeny derived by using oligonucleotide characters 1–50. Some of these were reported earlier but assigned different numbers (6). Five additional oligonucleotide characters, 51–55, were confined to single taxa and thus were not informative. All but one node were defined by multiple oligonucleotides or those that embraced multiple base changes or deletions. The one node defined by a single “one-base-change” oligonucleotide, 42, terminated a branch that contained Rattus satarae and two clusters of taxa that each were supported by multiple oligonucleotide characters. Is the hybridization of oligonucleotide 42 to R. satarae a case of parallelism; i.e., the independent amplification in R. satarae of an L1 subfamily that hybridizes to oligonucleotide 42? This oligonucleotide recognizes the change of an “A” (the ancestral base) to a “T” at position 10 in a stretch of 19 bases. The probability of this occurring by chance (no biological constraints) would be the product of the probabilities of position 10 changing from an “A” to “T” and of positions 1–9 and 11–19 not changing divided by 19. Calculating such probabilities would involve both numerous assumptions and knowledge of the mutation rate for L1, which is not known.
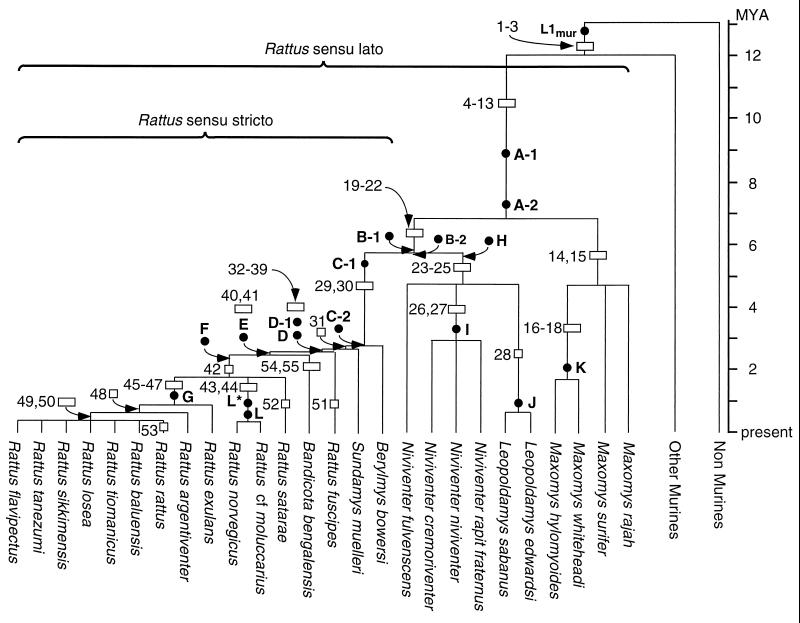
Figure 2.
The phylogeny of rats. This tree was built by using shared L1 oligonucleotide characters such as those numbered in Fig. 1. An open rectangle signifies that more than one oligonucleotide defines the branch, and an open square indicates that only one oligonucleotide character was used. The position of the oligonucleotide characters on a branch is arbitrary and not related to the “age” of the phylogenetic character. The length of the branches or positions of nodes in My was estimated, where possible, from the age of the indicated L1 subfamilies as described in the text. These subfamilies are a subset those in Fig. 1 and are positioned on the tree according to their age (solid circles). When no L1 subfamily was available to date a node or estimate a branch length they were drawn arbitrarily.
Therefore, we estimated the probability of the chance appearance of the type of sequence detected by oligonucleotide 42 (i.e., a 19-mer with a single change at position 10 to a “T”) from the frequency of such “one-base-change” 19-mers in our entire data set. We counted 19 (0.18%) occurrences of an “A” (ancestral base) to a “T” and 69 (0.64%) of any base to a “T” anywhere in our data set of ≈11,000 possible 19-mers. We only counted changes to one base (and not all possible bases) because this is what we detect with a single “one-base-change” oligonucleotide. We used the change to “T” because that is the change we scored, and in any case the frequency of 19-mers with a single change to each of the other bases was lower. Therefore, since the frequency of a character as the one detected by oligonucleotide 42 (a single-base change at position 10 in a 19-mer) is less than 1% we suggest that there is a greater than 95% probability that the position R. satarae on the tree in Fig. 2 is correct.
Only one tree is consistent with our data, and, as will be elaborated in the Discussion, it both confirms our earlier conclusions on the branching pattern in Rattus sensu lato and greatly extends our understanding of the evolutionary history and relationships within Rattus sensu stricto. Although this tree was readily constructed “by hand” we also organized the data into a matrix (not shown) wherein the presence or absence of the 50 informative characters was assigned the value of 1 or 0, respectively. Maximum parsimony analysis (25) was carried out on the data set assuming that the absence of a particular L1 character corresponds to the ancestral state and its presence to the derived state. A tree of identical topology to that shown in Fig. 2 was produced with a consistency index (CI) equal to 1, which means there were no homoplasic characters in our data set.
We estimated the length of the branches of the phylogenetic tree by using the calculated age (see Materials and Methods) of a subset of the L1 subfamilies listed in Fig. 1. We used only those subfamilies that contained elements cloned from at least some of the most widely diverged genera descendant from the branch in question. For example, the A-1 and A-2 families included members cloned from various species of Maxomys and some of the most recently emerged Rattus species, e.g., Rattus flavipectus. We also used only the oldest and youngest (most divergent and least divergent, respectively) subfamilies that satisfied these criteria for the particular branch. Seventeen of the 45 L1 subfamilies listed in Fig. 1 met these criteria and are positioned on Fig. 2 according to their age. Of these, five were between ≈5.5 and ≈7.5 My old and five were tightly clustered at ≈2.7 My. This latter intense period of L1 evolution coincided with and defines an intense speciation event that gave rise to five lineages: B. bowersi, S. muelleri, R. fuscipes, B. bengalensis, and the lineage that eventually gave rise to R. satarae, (R. cf moluccarius/R. norvegicus), and the lineage that led to R. exulans, R. argentiventer, and the (R. rattus… R. flavipectus) group. As shown in Fig. 2, we included B. bowersi, S. muelleri, and B. bengalensis in the Rattus sensu stricto group. We justify this for two reasons: First, a traditional member of Rattus sensu stricto, R. fuscipes, clusters among the newly included taxa. Second, all of the above-mentioned taxa are closely clustered and well separated from the remaining members of Rattus sensu lato by the long branch defined by the C-1 and C-2 L1 subfamilies.
Our use of the divergence of an L1 subfamily to estimate its age assumes (i) that the neutral substitution rate is approximately the same in all of the rodents considered here, (ii) that most members of a given family were inserted over a time shorter than the total length of time the subfamily has resided in the genome; and (iii) that, since their insertion, most of the members of a given subfamily have been diverging as pseudogenes (i.e., at the neutral substitution rate). There is good evidence that the neutral substitution rate is similar in murine rodents (30), and the standard deviation for the age of particularly the older subfamilies suggests that the second assumption is quite reasonable. If most members of a subfamily are diverging from each other at the pseudogene rate, then the sequence divergence between the members of a given L1 subfamily should reflect the neutral substitution rate of their host genomes. About 90% or more of the nonrepeated (single-copy) DNA fraction of mammalian genomes is thought to serve no coding function (e.g., see ref. 31) and thus presumably is not under selective pressure. Therefore, the divergence between the single-copy DNA fraction of mammalian genomes should largely reflect the neutral mutation rate. This divergence has been determined accurately for certain rodent genomes by DNA/DNA hybridization, and such measurements have been used to both order and date certain rodent speciation events (32). Therefore, the speciation times calculated from the age of L1 subfamilies should agree with those estimated from the DNA/DNA hybridization data.
In Table 1 we compare the times when the various taxa split from each other estimated from the age (divergence) of L1 subfamilies to those calculated by Ruedas and Kirsch from their DNA/DNA hybridization data (33). Row 1 shows that the age of the L1mur subfamily and the DNA/DNA hybridization data gave about the same time for the split of Rattus sensu lato from other murines. In Row 2, the DNA/DNA hybridization indicates that Maxomys split from the rest of Rattus sensu lato ≈7.6 Mya. The youngest L1 subfamily that we have identified that is still shared between Maxomys and the rest of Rattus sensu lato is the ≈7.3-My-old A-2 subfamily, and the oldest L1 subfamily identified so far in Rattus sensu lato not present in Maxomys is the ≈5.7-My-old B-1 subfamily. Therefore, we conclude that Maxomys split from the other Rattus sensu lato taxa sometime between ≈7.3 and ≈5.7 Mya, consistent with the estimate from the DNA/DNA hybridization data. The rest of Table 1 shows good agreement between the two methods and indicates that the age (divergence) of L1 subfamilies can be used to estimate branch lengths for phylogenetic trees built on the L1 character.
Table 1.
Comparison of the divergence times (in My) of various rodents estimated from the age of L1 subfamilies with those derived from DNA/DNA hybridization
| Dichotomy compared | L1 subfamily(ies) Time (or range) of split | DNA/DNA hybridization | |
|---|---|---|---|
| Rattus sensu lato/ | L1mur1* | ||
| other Murines | 12 ± 2.5 | 12.2† | |
| Maxomys/other rats | A-2 | B-1 | |
| <7.3 ± 1.4 | >5.7 ± 1.6 | 7.6 | |
| Maxomys rajah/ | A-2 | K | |
| M. whiteheadi | <7.3 ± 1.4 | >2.1 ± 0.7 | 4.3 |
| Leopoldamys and | H | ||
| Niviventer/other rats | ≈5.7 ± 0.7 | 5.4 | |
| Leopoldamys/Niviventer | B-2 | I | |
| <5.3 ± 1.4 | >3.4 ± 0.7 | 3.3 | |
| Berylmys/other rats | C-2 | ||
| ≈2.8 ± 1.3‡ | 3.3 | ||
| Sundamys/other rats | D-1 | ||
| ≈2.9 ± 1.1 | 2.5 | ||
| Bandicota/other rats | F | ||
| ≈2.6 ± 0.9 | 2.5 | ||
DISCUSSION
Here we significantly advanced the phylogeny of Rattus, which can serve as a framework for comparative morphology and traditional classification. In doing so we demonstrated the robust and unique properties of L1 as a phylogenetic character that can both determine and date speciation events. We expanded the definition of the Rattus sensu stricto group to include Berylmys, Sundamys, and Bandicota; resolved heretofore unknown branching patterns within the Rattus sensu stricto group; and demonstrated that the speciation events that both generated and further differentiated the Rattus sensu stricto lineage were both episodic and quite intense.
Five L1 subfamilies (A-2, B-1, B-2, H, and C-1) arose and amplified from ≈7.3 Mya to ≈5.4 Mya (Figs. 1 and 2). During this time the lineages for Maxomys, the (Niviventer/Leopoldamys) group, and Rattus sensu stricto were established. After ≈3 My of apparent stasis, an episode of intense speciation occurred in Rattus sensu stricto and generated Berylmys, Sundamys, R. fuscipes, Bandicota, and the lineage that eventually gave rise to R. satarae, the (R. cf moluccarius/R. norvegicus) group, and the (R. exulans… R. flavipectus) group. Coincident with this speciation was a period of intense L1 evolution that generated five L1 subfamilies (C-2, D, D-1, E, and F) in less than ≈0.3 My, which permitted the ordering and dating of this near simultaneous speciation event. Therefore, the L1 phylogenetic character evolved and amplified rapidly enough to generate unambiguous phylogenetic signals even when the nodes of the tree are joined by extremely short branches. Thus, as with SINE insertion events (9, 34, 35), amplified novel L1 subfamilies can be considered near ideal cladistic characters; the ancestral state of the character is never an issue, and the likelihood of parallelism, reversion, or convergence is most unlikely. However, in contrast to SINE insertions, the age of the L1 character (subfamily) can be estimated and used to date speciation events.
Our results on the above speciation episodes agree with previous DNA/DNA hybridization studies on the relationship between the Maxomys, Niviventer, Leopoldamys, and Rattus sensu stricto (Fig. 2, Table 1, and ref. 33). The DNA/DNA hybridization results also demonstrated that Berylmys, Sundamys, and Bandicota are each closely related to other taxa of Rattus sensu stricto (Fig. 2, Table 1, and refs. 33 and 36). The latter study showed, as we found, that Sundamys is closer to Rattus sensu stricto than Berylmys but they inferred a branching pattern of (Bandicota (Sundamys, Rattus)) instead of our (Sundamys (Bandicota, Rattus)). A close relationship between Bandicota and Rattus sensu stricto also was inferred by others (e.g., ref. 15) who used isozyme data to place Bandicota in Rattus sensu stricto as we did here. Chevret (36) also proposed a close relationship between Bandicota and Rattus sensu stricto based on DNA/DNA hybridization and suggested that Bandicota split from Rattus sensu stricto ≈2 Mya, which agrees with our timing (≈2.5 My, subfamily E) for this split (cf. Figs. 1 and 2). However, neither the DNA/DNA hybridization study (36) nor any other previous study resolved any of the branches that we found after Bandicota diverged and found no evidence for the second wave of speciation that we detected within Rattus sensu stricto. As Fig. 2 shows, this began ≈1.2 Mya and is marked by the emergence and amplification of L1 subfamily G (Figs. 1 and 2).
Despite our efforts, we have not yet found L1 characters to resolve branching within the seven taxon cluster of (R. rattus… R. flavipectus). Altogether we sequenced 91 elements from these taxa and used L1 oligonucleotide probes defined on both distinctive regions of individual L1 elements as well as on diagnostic regions of L1 subfamilies. That we found L1 subfamilies young enough to mark the split between R. norvegicus and R. cf moluccarius, which occurred about 0.5 Mya (5), and have identified a 0.3-My-old L1 subfamily (L1rat360) in R. satarae means that L1 elements amplify fast enough to theoretically examine events at least as old as several hundred thousand years. Perhaps we merely have been unlucky in our hunt for phylogenetically informative L1 elements in the seven-taxon cluster. Our identification of an L1 oligonucleotide character (53) confined to one member of this cluster, R. rattus (Fig. 2), indicates that our sample was large enough to reveal at least some distinctive L1 elements within the seven-taxon cluster. Therefore, perhaps our failure to identify L1 characters that define a branching pattern between these taxa means that they cannot be related by a tree because each is an independent lineage from a single archaic rat taxon.
One question raised by our results is whether there is any evidence for the three episodes of Rattus speciation in the fossil record? Unfortunately, fossils of rat-like murines for the period from 10 to 3 Mya (Upper Miocene and Middle and Lower Pliocene) that could critically address the two older speciation events of ≈7.3 to ≈5.7 Mya and ≈2.7 Mya have yet to be found. However, dating of the fossils that have been discovered so far is consistent with our conclusions. For example, Fig. 2 shows that the lineages for Maxomys, Rattus sensu stricto, and the (Niviventer/Leopoldamys) group diverged ≈7.3 to ≈5.7 Mya. Therefore, fossils of each of these lineages should be present at sites dated after this time, and such fossils have been recovered from sites dated from ≈3 to ≈1.8 Mya in both Thailand (37, 38) and China (39). Furthermore, Fig. 2 shows that speciation within Niviventer and Leopoldamys started after ≈3.5 Mya and fossils for each genus have been recovered from sites dated at ≈2.0 Mya in China (39). And finally, Fig. 2 shows that Berylmys and Sundamys emerged ≈3 Mya. The discovery of fossils for these genera in deposits that dated at ≈2 Mya (38, 40, 41) indicate that these lineages were established before then. However, in contrast to the two older speciation episodes, the one that we detected within Rattus sensu stricto beginning at ≈1.2 Mya is well supported by the fossil record that shows increased speciation within Rattus beginning ≈1.6 Mya (the Middle Pleistocene; refs. 37 and 39).
Our proposed phylogeny for Rattus sensu stricto includes episodes of intense speciation that we could detect and partially resolve because of the rapidity with which L1 elements evolve and amplify. These results have important implications for the dynamics and mechanism of evolution of both L1 elements and their rodent hosts. For example, the near simultaneous generation of at least five rat lineages ≈2.7 Mya is a paleobiological event that may reflect the population structure of the ancient rodent population and paleogeographical or paleoenvironmental events of Southeast Asia whence these species originated. We would also expect that using L1 phylogenetic characters to address phylogenetic and evolutionary questions in other mammalian taxa also may reveal details of their evolutionary history not readily addressed by current methods. We have found, in studies to be reported elsewhere (S. Boissinot, P. Chevret, and A.V.F., unpublished data), that relationships within primates and within and between Old World and New World monkeys can be determined readily by using L1 elements and are now investigating several questions regarding modern primate taxa. In particular, the rapidity with which novel L1 subfamilies appear and amplify suggests that this could be occurring in present-day taxa. Therefore, it is possible that populations within a species could be distinguished by their variant L1 subfamilies.
Acknowledgments
We thank Drs. Dan L. Sackett and Allen P. Minton (National Institutes of Health) for insightful discussions and advice. O.V. was supported in part by a grant from La Fondation Singer-Polignac, 43, Avenue Geoges-Mandel, Paris.
ABBREVIATIONS
- My
million years
- Mya
My ago
- UTR
untranslated region
- LINE
long interspersed repeated DNA element
- SINE
short interspersed repeated DNA element
- L1
LINE-1
Footnotes
References
- 1.Hennig W. Phylogenetic Systematics. Urbana: Univ. of Illinois Press; 1966. [Google Scholar]
- 2.Stewart C-B. Nature (London) 1993;361:603–607. doi: 10.1038/361603a0. [DOI] [PubMed] [Google Scholar]
- 3.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 4.Furano A V, Usdin K. J Biol Chem. 1995;270:25301–24304. doi: 10.1074/jbc.270.43.25301. [DOI] [PubMed] [Google Scholar]
- 5.Usdin K, Chevret P, Catzeflis F M, Verona R, Furano A V. Mol Biol Evol. 1995;12:73–82. doi: 10.1093/oxfordjournals.molbev.a040192. [DOI] [PubMed] [Google Scholar]
- 6.Verneau O, Catzeflis F, Furano A V. J Mol Evol. 1997;45:424–436. doi: 10.1007/pl00006247. [DOI] [PubMed] [Google Scholar]
- 7.Batzer M A, Stoneking M, Alegria-Hartman M, Bazan H, Kass D H, Shaikh T H, Novick G E, Ioannou P A, Scheer W D, Herrera R J, Deininger P L. Proc Natl Acad Sci USA. 1994;91:12288–12292. doi: 10.1073/pnas.91.25.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murata S, Takasaki N, Saitoh M, Tachida H, Okada N. Genetics. 1996;142:915–926. doi: 10.1093/genetics/142.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimamura M, Yasue H, Ohshima K, Abe H, Kato H, Kishiro T, Goto M, Munechika I, Okada N. Nature (London) 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Terai Y, Nishida M, Okada N. Mol Biol Evol. 1998;15:391–407. doi: 10.1093/oxfordjournals.molbev.a025936. [DOI] [PubMed] [Google Scholar]
- 11.Serdobova I M, Kramerov D A. J Mol Evol. 1998;46:202–214. doi: 10.1007/pl00006295. [DOI] [PubMed] [Google Scholar]
- 12.Cook J M, Tristem M. Trends Ecol Evol. 1997;12:295–297. doi: 10.1016/S0169-5347(97)01121-X. [DOI] [PubMed] [Google Scholar]
- 13.Chan K L, Dhaliwal S S, Yong H S. Comp Biochem Physiol. 1979;64B:329–337. doi: 10.1016/0305-0491(79)90278-5. [DOI] [PubMed] [Google Scholar]
- 14.Pasteur N, Worms J, Tohari M, Iskandar D. Biochem Syst Ecol. 1982;10:191–196. [Google Scholar]
- 15.Gemmeke H, Niethammer J. Z Säugetierkunde. 1984;49:104–116. [Google Scholar]
- 16.Baverstock P R, Adams M, Watts C H S. Genetica. 1986;71:11–22. [Google Scholar]
- 17.Catzeflis F. Trends Ecol Evol. 1991;6:168. doi: 10.1016/0169-5347(91)90060-B. [DOI] [PubMed] [Google Scholar]
- 18.Musser G G, Carleton M D. In: Mammal Species of the World: A Taxonomic and Geographic Reference. 2nd Ed. Wilson D E, Reeder D M, editors. Washington, D.C./London: Smithsonian Inst. Press; 1993. pp. 501–755. [Google Scholar]
- 19.Hayward B E, Zavanelli M, Furano A V. Genetics. 1997;146:641–654. doi: 10.1093/genetics/146.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A. Current Protocols in Molecular Biology. New York: Wiley; 1989. [Google Scholar]
- 21.Buluwela L, Forster A, Boehm T, Rabbitts T H. Nucleic Acids Res. 1989;17:452. doi: 10.1093/nar/17.1.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philippe H. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1993. , Version 3.1.1. [Google Scholar]
- 26.Kimura M. Proc Natl Acad Sci USA. 1981;78:454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W-H, Tanimura M, Sharp P M. J Mol Evol. 1987;25:330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- 28.Pascale E, Liu C, Valle E, Usdin K, Furano A V. J Mol Evol. 1993;36:9–20. doi: 10.1007/BF02407302. [DOI] [PubMed] [Google Scholar]
- 29.Furano A V, Hayward B E, Chevret P, Catzeflis F, Usdin K. J Mol Evol. 1994;38:18–27. doi: 10.1007/BF00175491. [DOI] [PubMed] [Google Scholar]
- 30.O’hUigin C, Li W-H. J Mol Evol. 1992;35:377–384. doi: 10.1007/BF00171816. [DOI] [PubMed] [Google Scholar]
- 31.Sutcliffe J G. Annu Rev Neurosci. 1988;11:157–198. doi: 10.1146/annurev.ne.11.030188.001105. [DOI] [PubMed] [Google Scholar]
- 32.Catzeflis F M, Dickerman A W, Michaux J, Kirsch J A W. In: Mammal Phylogeny: Placentals. Szalay F S, Novacek M J, McKenna M C, editors. Vol. 2. New York: Springer; 1993. pp. 159–172. [Google Scholar]
- 33.Ruedas L A, Kirsch J A W. Biol J Linn Soc London. 1997;61:385–408. [Google Scholar]
- 34.Takasaki N, Park L, Kaeriyama M, Gharrett A J, Okada N. J Mol Evol. 1996;42:103–116. doi: 10.1007/BF02198835. [DOI] [PubMed] [Google Scholar]
- 35.Takasaki N, Yamaki T, Hamada M, Park L, Okada N. Genetics. 1997;146:369–380. doi: 10.1093/genetics/146.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chevret P. Ph.D. Thesis. Montpellier, France: Univ. of Montpellier II; 1994. [Google Scholar]
- 37.Chaimanee Y, Suteethorn V, Triamwichanon S, Jaeger J-J. C R Acad Sci Ser IIa. 1996;322:155–162. [Google Scholar]
- 38.Chaimanee Y. Les Rongeurs du Plio-Pleistocene de Thailande. Montpellier, France: Université Montpellier 2; 1997. [Google Scholar]
- 39.Zheng S. Quartenary Rodents of Sichuan-Guizhou Area, China. Peking: Science Press; 1993. [Google Scholar]
- 40.Medway L. Sarawak Museum J. 1964;11:616–623. [Google Scholar]
- 41.Medway L. In: Transactions of the Second Aberdeen-Hull Symposium on Malesian Ecology. Ashton P, Ashton M, editors. Vol. 13. Hull, U.K.: Univ. of Hull; 1972. pp. 63–98. [Google Scholar]
- 42.Catzeflis F M, Aguilar J-P, Jaeger J-J. Trends Ecol Evol. 1992;7:122–126. doi: 10.1016/0169-5347(92)90146-3. [DOI] [PubMed] [Google Scholar]