Abstract
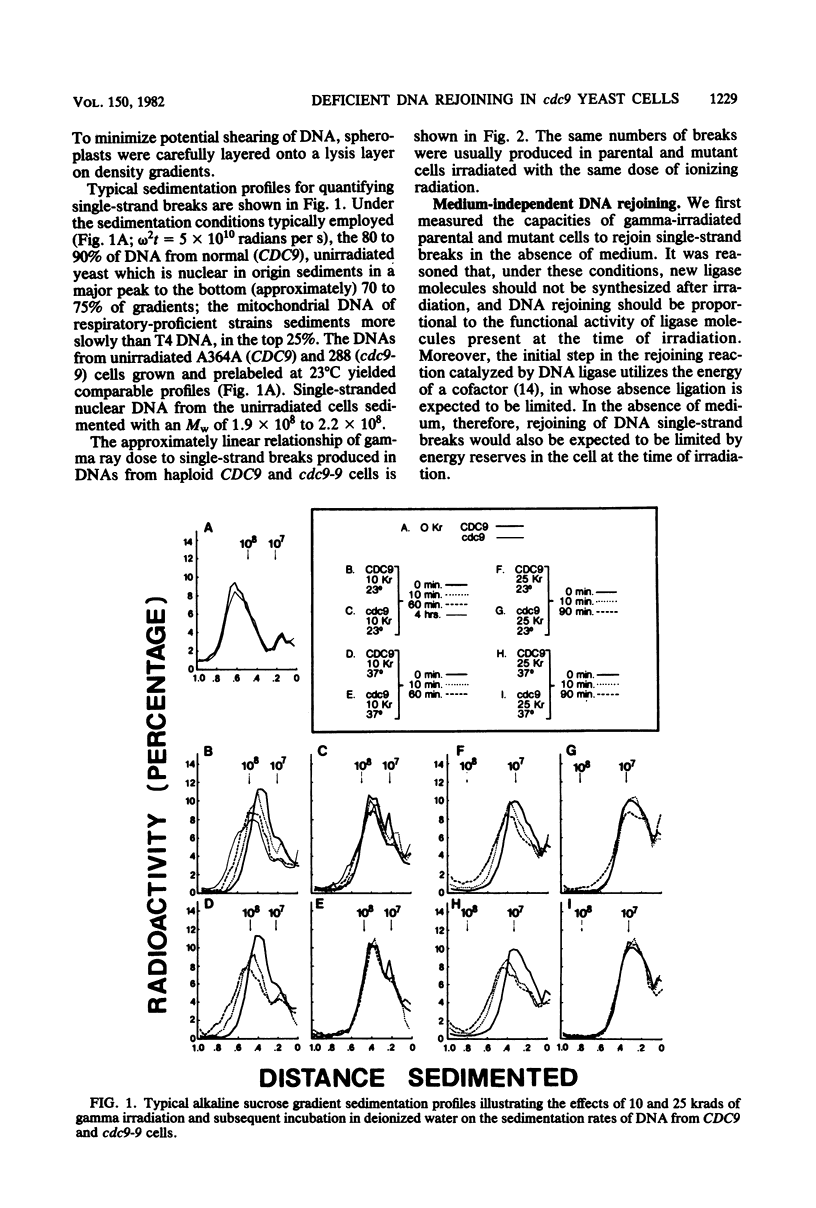
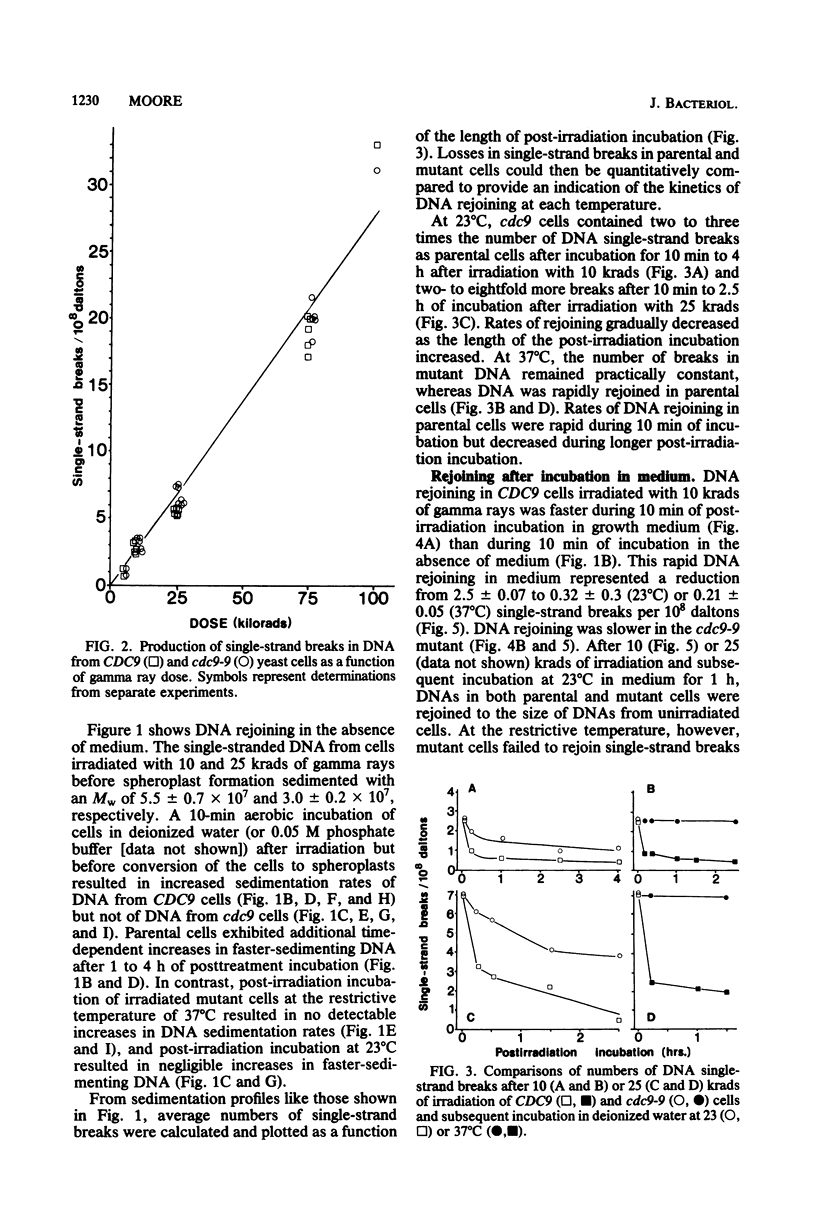
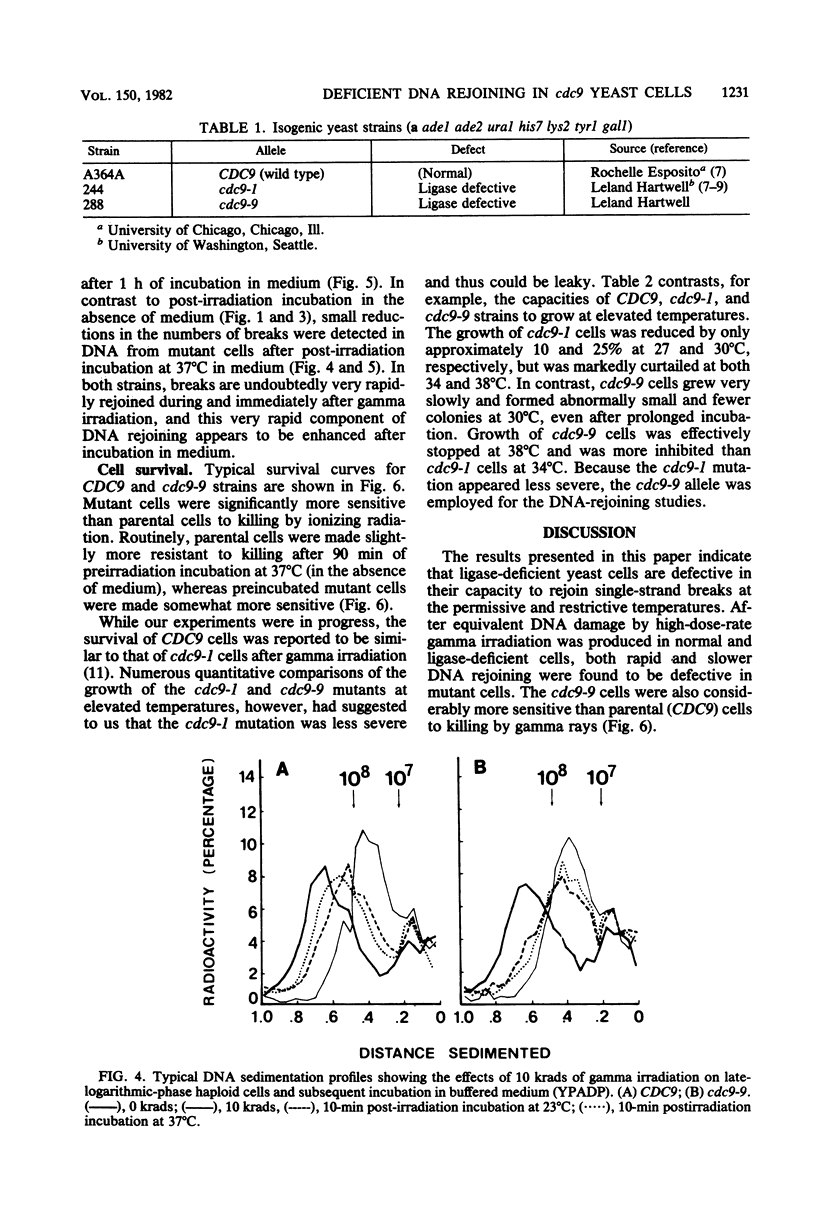
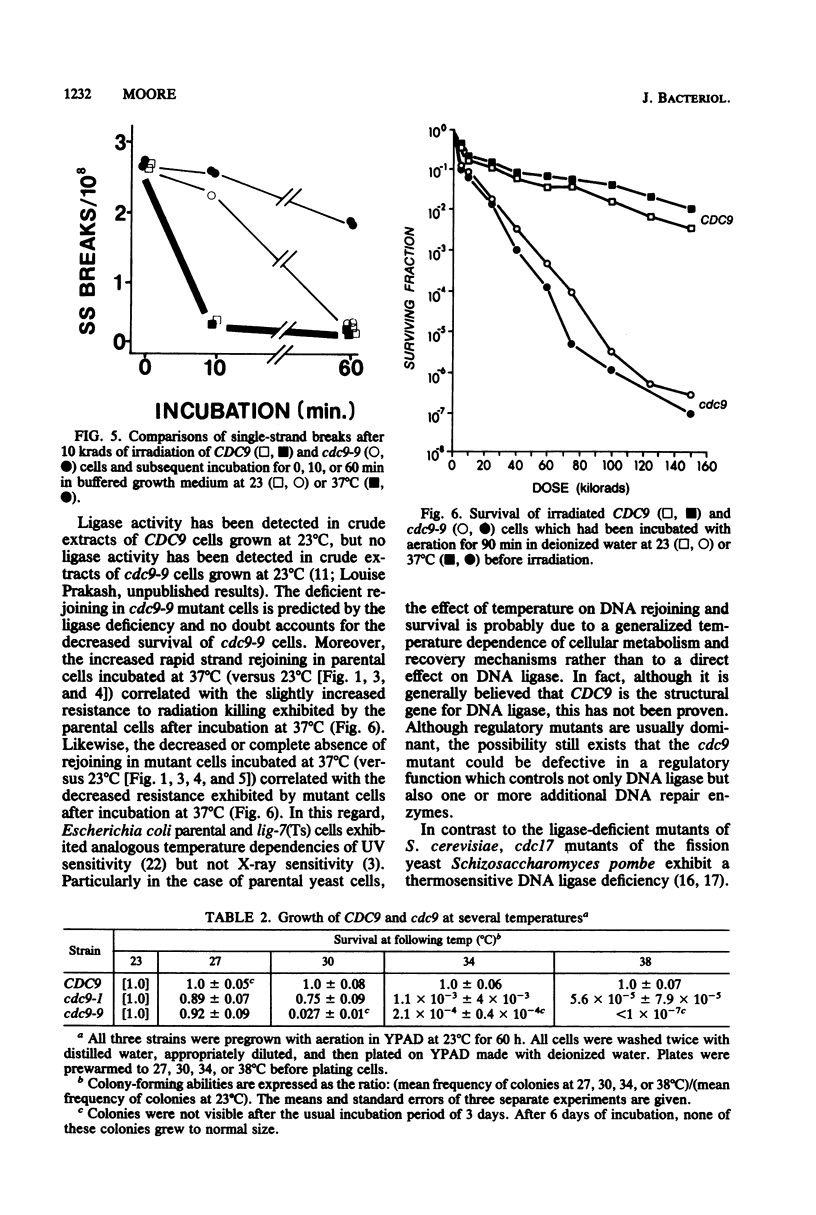
Yeast cells deficient in DNA ligase were also deficient in their capacity to rejoin single-strand scissions in prelabeled nuclear DNA. After high-dose-rate gamma irradiation (10 and 25 krads), cdc9-9 mutant cells failed to rejoin single-strand scissions at the restrictive temperature of 37 degrees C. In contrast, parental (CDC9) cells (incubated with mutant cells both during and after irradiation) exhibited rapid medium-independent DNA rejoining after 10 min of post-irradiation incubation and slower rates of rejoining after longer incubation. Parental cells were also more resistant than mutant cells to killing by gamma irradiation. Approximately 2.5 +/- 0.07 and 5.7 +/- 0.6 single-strand breaks per 10(8) daltons were detected in DNAs from either CDC9 or cdc9-9 cells converted to spheroplasts immediately after 10 and 25 krads of irradiation, respectively. At the permissive temperature of 23 degrees C, the cdc9-9 cells contained 2 to 3 times the number of DNA single-strand breaks as parental cells after 10 min to 4 h of incubation after 10 krads of irradiation, and two- to eightfold more breaks after 10 min to 2.5 h of incubation after 25 krads of irradiation. Rejoining of single-strand scissions was faster in medium. After only 10 min in buffered growth medium and after 10 krads of irradiation, the number of DNA single-strand breaks was reduced to 0.32 +/- 0.3 (at 23 degrees C) or 0.21 +/- 0.05 (at 37 degrees C) per 10(8) daltons in parental cells, but remained at 2.1 +/- 0.06 (at 23 degrees C) or 2.3 +/- 0.07 (at 37 degrees C) per 10(8) daltons in mutant cells. After 10 or 25 krads of irradiation plus 1 h of incubation in medium at 37 degrees C, only DNA from CDC9 cells was rejoined to the size of DNA from unirradiated cells, whereas at 23 degrees C, DNAs in both strains were completely rejoined.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carrier W. L., Setlow R. B. Paper strip method for assaying gradient fractions containing radioactive macromolecules. Anal Biochem. 1971 Oct;43(2):427–432. doi: 10.1016/0003-2697(71)90272-7. [DOI] [PubMed] [Google Scholar]
- Fabre F., Roman H. Evidence that a single DNA ligase is involved in replication and recombination in yeast. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4586–4588. doi: 10.1073/pnas.76.9.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte M. A., Fangman W. L. Naturally occurring cross-links in yeast chromosomal DNA. Cell. 1976 Jul;8(3):425–431. doi: 10.1016/0092-8674(76)90155-0. [DOI] [PubMed] [Google Scholar]
- Game J. C., Johnston L. H., von Borstel R. C. Enhanced mitotic recombination in a ligase-defective mutant of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4589–4592. doi: 10.1073/pnas.76.9.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976 Jul 15;104(4):803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Horiuchi T., Sato T., Nagata T. DNA degradation in an amber mutant of Escherichia coli K12 affecting DNA ligase and viability. J Mol Biol. 1975 Jun 25;95(2):271–287. doi: 10.1016/0022-2836(75)90395-2. [DOI] [PubMed] [Google Scholar]
- Johnston L. H., Nasmyth K. A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978 Aug 31;274(5674):891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- Johnston L. H. The DNA repair capability of cdc9, the Saccharomyces cerevisiae mutant defective in DNA ligase. Mol Gen Genet. 1979 Feb 16;170(1):89–92. doi: 10.1007/BF00268583. [DOI] [PubMed] [Google Scholar]
- Konrad E. B., Modrich P., Lehman I. R. Genetic and enzymatic characterization of a conditional lethal mutant of Escherichia coli K12 with a temperature-sensitive DNA ligase. J Mol Biol. 1973 Jul 15;77(4):519–529. doi: 10.1016/0022-2836(73)90220-9. [DOI] [PubMed] [Google Scholar]
- Lehman I. R. DNA ligase: structure, mechanism, and function. Science. 1974 Nov 29;186(4166):790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- Moore C. W., Vossler D. A. Influence of pH and length of post-treatment incubation on bleomycin-induced DNA damage. Biochim Biophys Acta. 1980 Dec 11;610(2):425–429. doi: 10.1016/0005-2787(80)90024-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A. Temperature-sensitive lethal mutants in the structural gene for DNA ligase in the yeast Schizosaccharomyces pombe. Cell. 1977 Dec;12(4):1109–1120. doi: 10.1016/0092-8674(77)90173-8. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Fangman W. L. Sedimentation properties of yeast chromosomal DNA. Proc Natl Acad Sci U S A. 1972 May;69(5):1188–1191. doi: 10.1073/pnas.69.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]



