Abstract
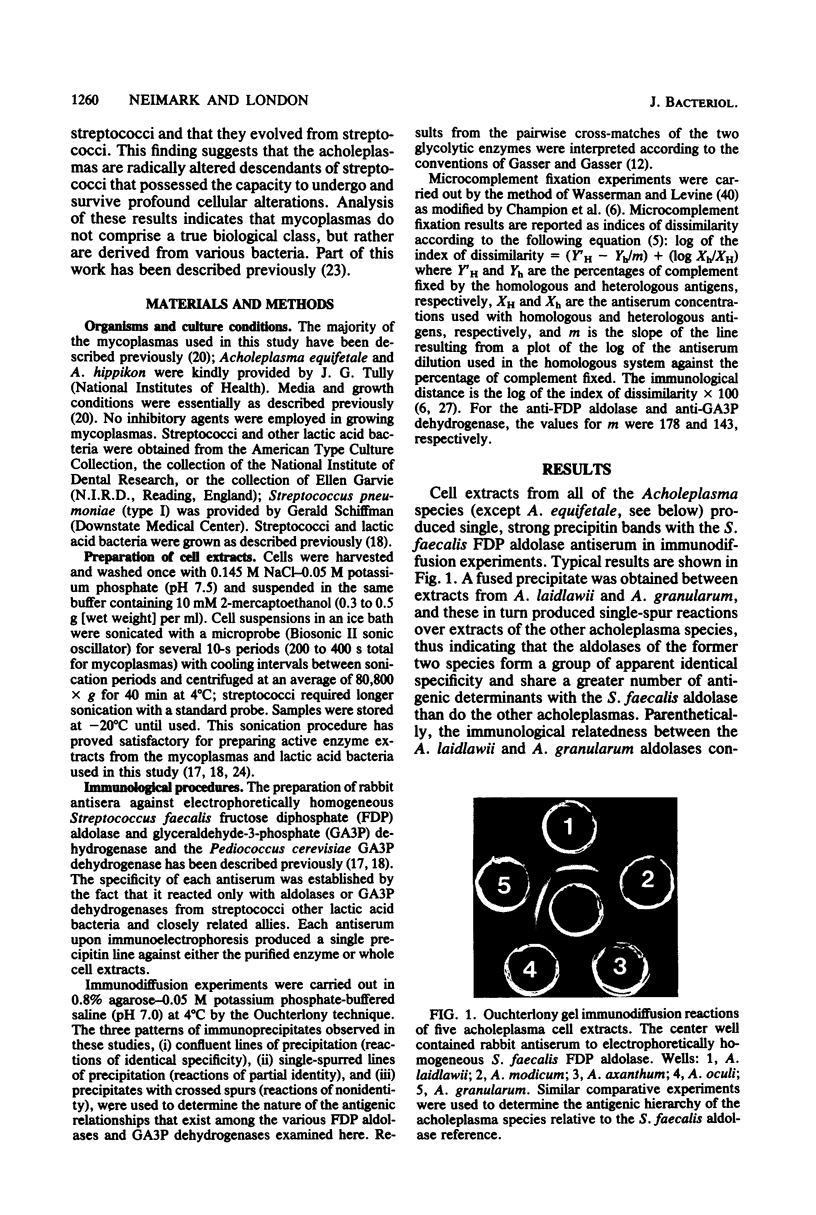
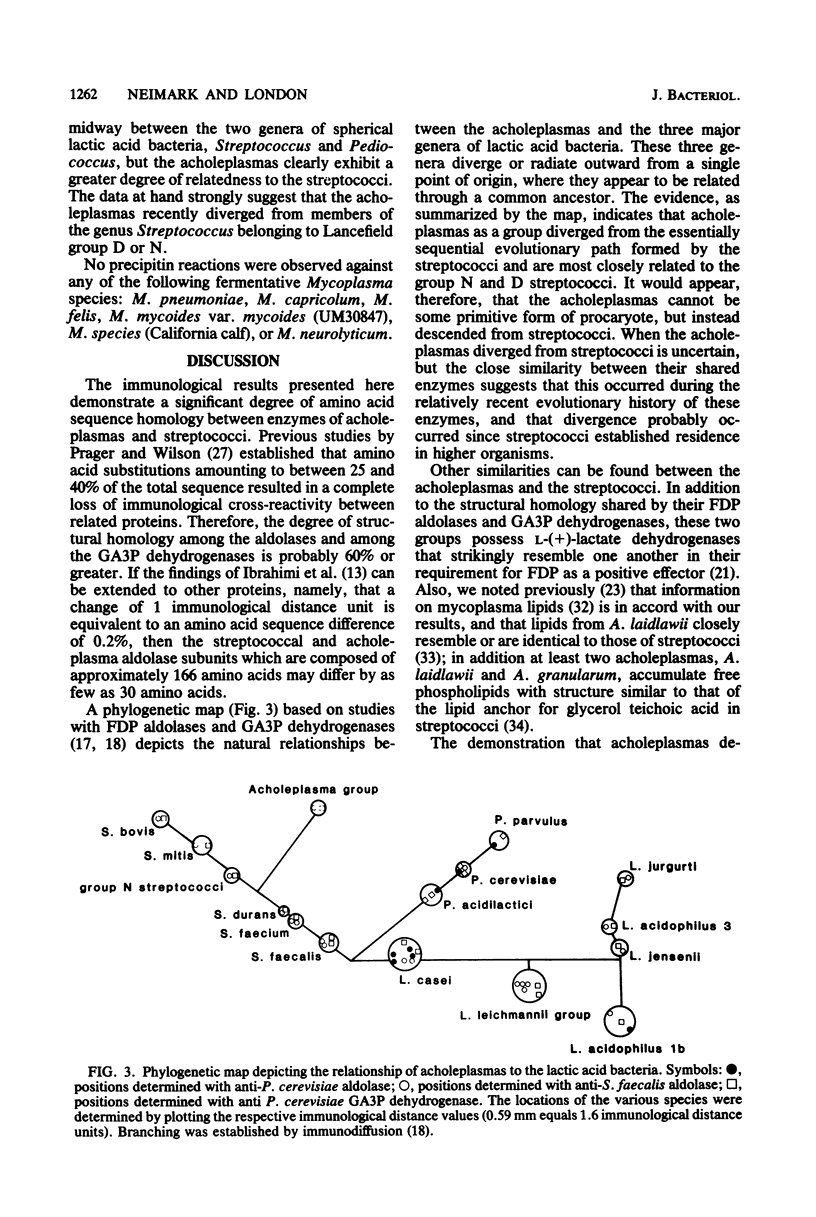
We report the establishment of a phylogenetic relationship between the sterol-nonrequiring mycoplasmas (Acholeplasma species) and streptococci. Three specific antisera prepared against purified Streptococcus faecalis fructose diphosphate aldolase and glyceraldehyde-3-phosphate dehydrogenase and Pediococcus cerevisiae glyceraldehyde-3-phosphate dehydrogenase were used for comparative enzyme immunological studies; the Ouchterlony double-diffusion technique and the quantitative microcomplement fixation procedure were employed. The reactions obtained provide evidence showing that all seven ACholeplasma species studied (A. laidlawii, A. granularum, A. modicum, A. oculi, A. axanthum. A. hippikon, and A. equifetale) are phylogenetically related to streptococci and that they evolved from streptococci. The data strongly suggest that the acholeplasmas comprise a distinct evolutionary group that has diverged from streptococci belonging to Lancefield group D or N. No reactions were observed between these enzyme antisera and cell extracts from six fermentative Mycoplasma species. These results support the view that mycoplasmas are derived from various bacteria.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Plasmids, loss of lactose metabolism, and appearance of partial and full lactose-fermenting revertants in Streptococcus cremoris B1. J Bacteriol. 1977 Jan;129(1):367–377. doi: 10.1128/jb.129.1.367-377.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak A. L., Black F. T., Christiansen C., Freundt E. A. Genome size of mycoplasmal DNA. Nature. 1969 Dec 20;224(5225):1209–1210. doi: 10.1038/2241209a0. [DOI] [PubMed] [Google Scholar]
- Bak A. L., Christiansen C., Stenderup A. Bacterial genome sizes determined by DNA renaturation studies. J Gen Microbiol. 1970 Dec;64(3):377–380. doi: 10.1099/00221287-64-3-377. [DOI] [PubMed] [Google Scholar]
- DIENES L., WEINBERGER H. J. The L forms of bacteria. Bacteriol Rev. 1951 Dec;15(4):245–288. doi: 10.1128/br.15.4.245-288.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDWARD D. G. Biology of the pleuropneumonialike organisms. Introduction. Ann N Y Acad Sci. 1960 Jan 15;79:309–311. doi: 10.1111/j.1749-6632.1960.tb42692.x. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Gasser F., Gasser C. Immunological relationships among lactic dehydrogenases in the genera Lactobacillus and Leuconostoc. J Bacteriol. 1971 Apr;106(1):113–125. doi: 10.1128/jb.106.1.113-125.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi I. M., Prager E. M., White T. J., Wilson A. C. Amino acid sequence of California quail lysozyme. Effect of evolutionary substitutions on the antigenic structure of lysozyme. Biochemistry. 1979 Jun 26;18(13):2736–2744. doi: 10.1021/bi00580a008. [DOI] [PubMed] [Google Scholar]
- Kimball M. E., Szeto K. S., Soll D. The nucleotide sequence of phenylalanine tRNA from Mycoplasma sp. (Kid). Nucleic Acids Res. 1974 Dec;1(12):1721–1732. [PMC free article] [PubMed] [Google Scholar]
- London J., Chace N. M. Aldolases of the lactic acid bacteria. Demonstration of immunological relationships among eight genera of Gram positive bacteria using an anti-pediococcal aldolase serum. Arch Microbiol. 1976 Oct 11;110(1):121–128. doi: 10.1007/BF00416976. [DOI] [PubMed] [Google Scholar]
- London J., Kline K. Aldolase of lactic acid bacteria: a case history in the use of an enzyme as an evolutionary marker. Bacteriol Rev. 1973 Dec;37(4):453–478. doi: 10.1128/br.37.4.453-478.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEIMARK H. C., PICKETT M. J. Products of glucose metabolism by pleuropneumonialike organisms. Ann N Y Acad Sci. 1960 Jan 15;79:531–537. doi: 10.1111/j.1749-6632.1960.tb42719.x. [DOI] [PubMed] [Google Scholar]
- Neimark H. C. Division of mycoplasmas into subgroups. J Gen Microbiol. 1970 Oct;63(2):249–263. doi: 10.1099/00221287-63-2-249. [DOI] [PubMed] [Google Scholar]
- Neimark H., Lemcke R. M. Occurrence and properties of lactic dehydrogenases of fermentative mycoplasmas. J Bacteriol. 1972 Sep;111(3):633–640. doi: 10.1128/jb.111.3.633-640.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neimark H., Tung M. C. Properties of a fructose-1,6-diphosphate-activated lactate dehydrogenase from Acholeplasma laidlawii type A. J Bacteriol. 1973 Jun;114(3):1025–1033. doi: 10.1128/jb.114.3.1025-1033.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager E. M., Wilson A. C. The dependence of immunological cross-reactivity upon sequence resemblance among lysozymes. II. Comparison of precipitin and micro-complement fixation results. J Biol Chem. 1971 Nov 25;246(22):7010–7017. [PubMed] [Google Scholar]
- RODWELL A. W. Nutrition and metabolism of Mycoplasma mycoides var. mycoides. Ann N Y Acad Sci. 1960 Jan 15;79:499–507. doi: 10.1111/j.1749-6632.1960.tb42716.x. [DOI] [PubMed] [Google Scholar]
- Razin S. The mycoplasmas. Microbiol Rev. 1978 Jun;42(2):414–470. doi: 10.1128/mr.42.2.414-470.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M., Anilionis A. Evolution of the bacterial genome. Annu Rev Microbiol. 1978;32:519–560. doi: 10.1146/annurev.mi.32.100178.002511. [DOI] [PubMed] [Google Scholar]
- SMITH S. L., VANDEMARK P. J., FABRICANT J. RESPIRATORY PATHWAYS IN THE MYCOPLASMA. I. LACTATE OXIDATION BY MYCOPLASMA GALLISEPTICUM. J Bacteriol. 1963 Nov;86:893–897. doi: 10.1128/jb.86.5.893-897.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw N. Lipid composition as a guide to the classification of bacteria. Adv Appl Microbiol. 1974;17(0):63–108. doi: 10.1016/s0065-2164(08)70555-0. [DOI] [PubMed] [Google Scholar]
- Smith P. F., Patel K. R., Al-Shammari A. J. An aldehydophosphoglycolipid from Acholeplasma granularum. Biochim Biophys Acta. 1980 Mar 21;617(3):419–429. doi: 10.1016/0005-2760(80)90008-9. [DOI] [PubMed] [Google Scholar]
- Starlinger P. IS elements and transposons. Plasmid. 1980 May;3(3):241–259. doi: 10.1016/0147-619x(80)90039-6. [DOI] [PubMed] [Google Scholar]
- Van Demark P. J. Respiratory pathways in the mycoplasma. Ann N Y Acad Sci. 1967 Jul 28;143(1):77–84. doi: 10.1111/j.1749-6632.1967.tb27647.x. [DOI] [PubMed] [Google Scholar]
- WASSERMAN E., LEVINE L. Quantitative micro-complement fixation and its use in the study of antigenic structure by specific antigen-antibody inhibition. J Immunol. 1961 Sep;87:290–295. [PubMed] [Google Scholar]
- Walker R. T., RajBhandary U. L. The nucleotide sequence of formylmethionine tRNA from Mycoplasma mycoides sp. capri. Nucleic Acids Res. 1978 Jan;5(1):57–70. doi: 10.1093/nar/5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Maniloff J., Zablen L. B. Phylogenetic analysis of the mycoplasmas. Proc Natl Acad Sci U S A. 1980 Jan;77(1):494–498. doi: 10.1073/pnas.77.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]





