Abstract
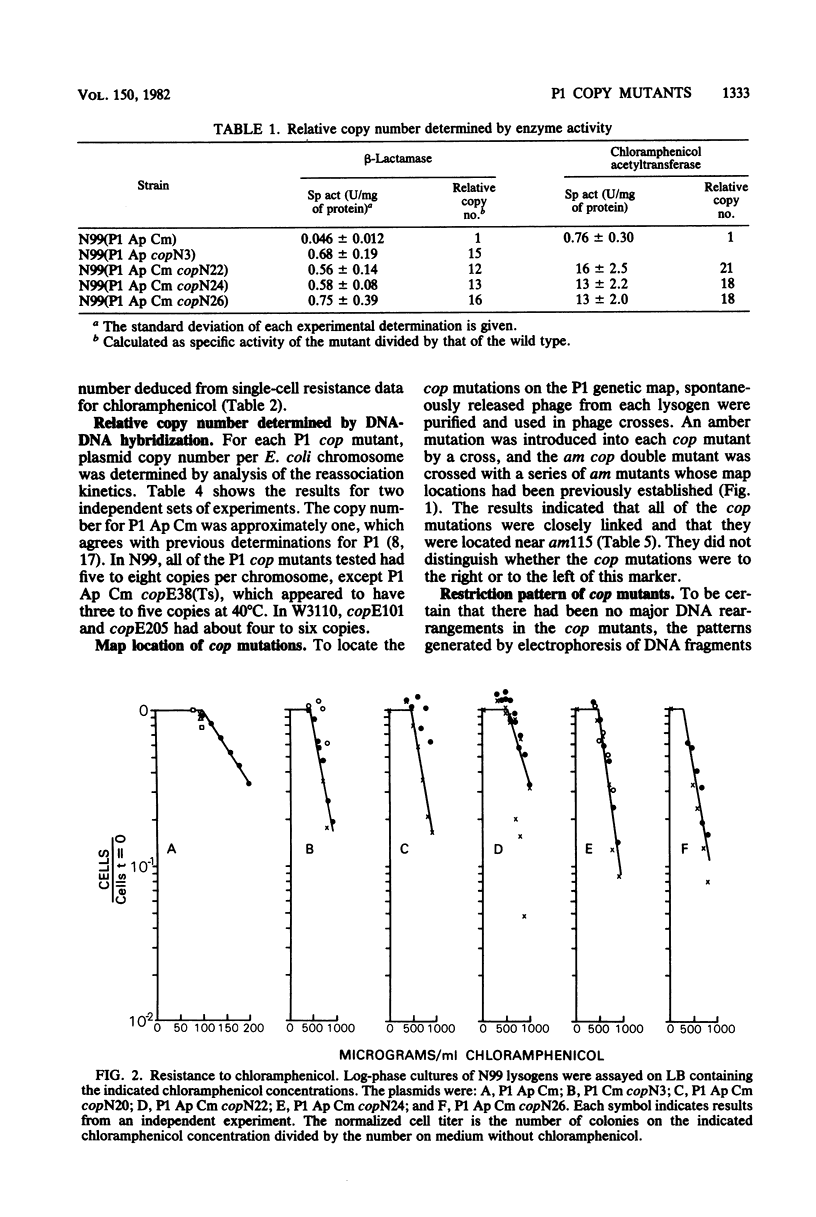
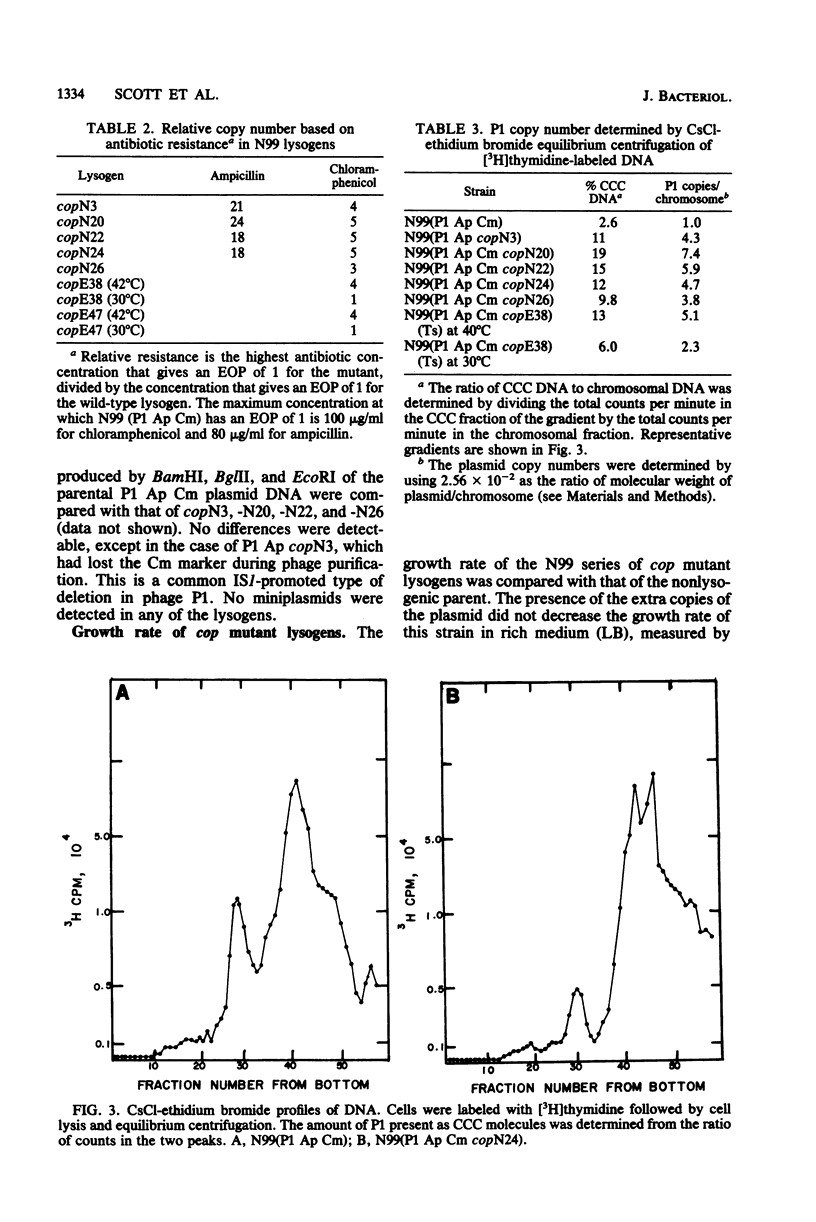
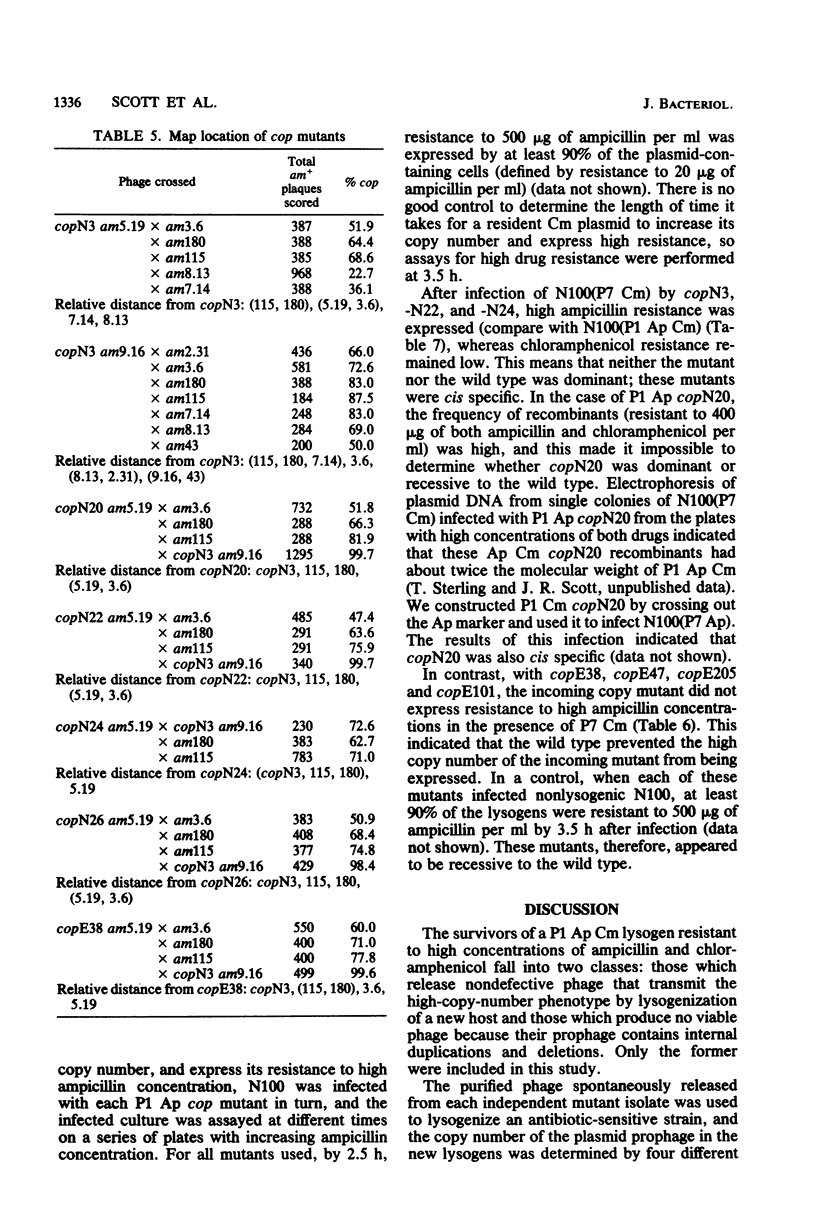
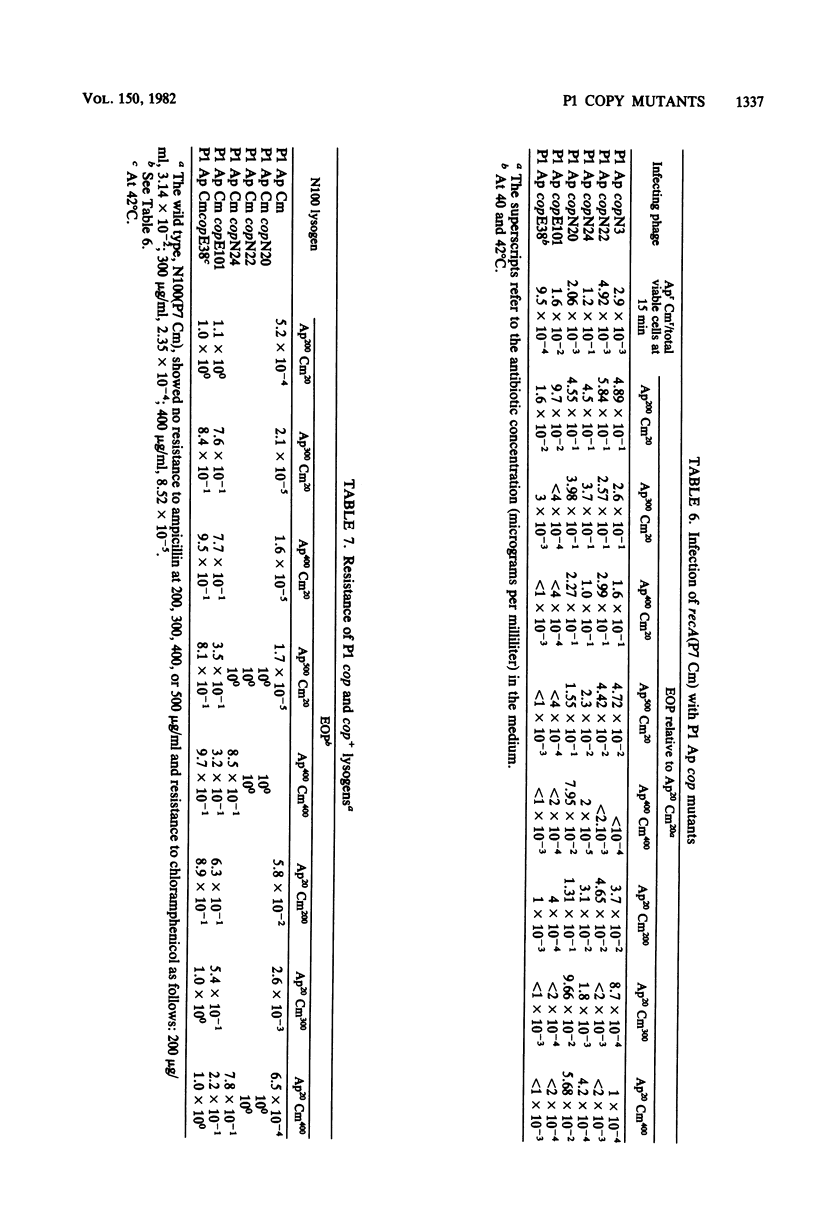
A series of mutations of the P1 plasmid prophage that lead to increased copy number was isolated and analyzed. The copy number of the mutants was elevated at least five- to eightfold relative to wild-type P1, as determined by single-cell resistance to antibiotics, activity of enzymes, content of superhelical DNA, and reassociation kinetics. The copy number of two of the mutants was temperature dependent. Based on dominance tests, the mutants fell into two classes, cis specific and recessive. The latter class included a temperature-sensitive copy mutant. The existence of a class of recessive mutants suggests that the replication of the P1 plasmid is negatively regulated.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bächi B., Arber W. Physical mapping of BglII, BamHI, EcoRI, HindIII and PstI restriction fragments of bacteriophage P1 DNA. Mol Gen Genet. 1977 Jun 24;153(3):311–324. doi: 10.1007/BF00431596. [DOI] [PubMed] [Google Scholar]
- Chesney R. H., Scott J. R. Suppression of a thermosensitive dnaA mutation of Escherichia coli by bacteriophage P1 and P7. Plasmid. 1978 Feb;1(2):145–163. doi: 10.1016/0147-619x(78)90035-5. [DOI] [PubMed] [Google Scholar]
- Chesney R. H., Scott J. R., Vapnek D. Integration of the plasmid prophages P1 and P7 into the chromosome of Escherichia coli. J Mol Biol. 1979 May 15;130(2):161–173. doi: 10.1016/0022-2836(79)90424-8. [DOI] [PubMed] [Google Scholar]
- Cowan J. A., Scott J. R. Incompatibility among group Y plasmids. Plasmid. 1981 Sep;6(2):202–221. doi: 10.1016/0147-619x(81)90067-6. [DOI] [PubMed] [Google Scholar]
- Davis R., Vapnek D. In vivo transcription of R-plasmid deoxyribonucleic acid in Escherichia coli strains with altered antibiotic resistance levels and/or conjugal proficiency. J Bacteriol. 1976 Mar;125(3):1148–1155. doi: 10.1128/jb.125.3.1148-1155.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb L. D., Kohne D. E., Martin M. A. Quantitation of Simian virus 40 sequences in African green monkey, mouse and virus-transformed cell genomes. J Mol Biol. 1971 Apr 14;57(1):129–145. doi: 10.1016/0022-2836(71)90123-9. [DOI] [PubMed] [Google Scholar]
- IMSANDE J. NEW ASSAY FOR PENICILLINASE AND SOME RESULTS ON PENICILLINASE INDUCTION. J Bacteriol. 1965 May;89:1322–1327. doi: 10.1128/jb.89.5.1322-1327.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. Prophage P1, and extrachromosomal replication unit. Cold Spring Harb Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- Jaffé-Brachet A., D'Ari R. Maintenance of bacteriophage P1 plasmid. J Virol. 1977 Sep;23(3):476–482. doi: 10.1128/jvi.23.3.476-482.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KONDO E., MITSUHASHI S. DRUG RESISTANCE OF ENTERIC BACTERIA. IV. ACTIVE TRANSDUCING BACTERIOPHAGE P1 CM PRODUCED BY THE COMBINATION OF R FACTOR WITH BACTERIOPHAGE P1. J Bacteriol. 1964 Nov;88:1266–1276. doi: 10.1128/jb.88.5.1266-1276.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lindqvist R. C., Nordström K. Resistance of Escherichia coli to penicillins. VII. Purification and characterization of a penicillinase mediated by the R factor R1. J Bacteriol. 1970 Jan;101(1):232–239. doi: 10.1128/jb.101.1.232-239.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell I. H., Van Ness J., Hahn W. E. Assay of DNA-RNA hybrids by S1 nuclease digestion and adsorption to DEAE-cellulose filters. Nucleic Acids Res. 1978 Jun;5(6):2033–2038. doi: 10.1093/nar/5.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Caro L., Berg C. M., Hirota Y. Chromosome replication in Escherichia coli. IV. Control of chromosome replication and cell division by an integrated episome. J Mol Biol. 1971 Feb 14;55(3):441–456. doi: 10.1016/0022-2836(71)90328-7. [DOI] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Caro L. Replication of prophage P1 during the cell cycle of Escherichia coli. Mol Gen Genet. 1977 Mar 28;152(1):71–76. doi: 10.1007/BF00264942. [DOI] [PubMed] [Google Scholar]
- Rawson J. R., Boerma C. L., Andrews W. H., Wilkerson C. G. Complexity and abundance of ribonucleic acid transcribed from restriction endonuclease fragments of Euglena chloroplast deoxyribonucleic acid during chloroplast development. Biochemistry. 1981 Apr 28;20(9):2639–2644. doi: 10.1021/bi00512a043. [DOI] [PubMed] [Google Scholar]
- Razza J. B., Watkins C. A., Scott J. R. Phage P1 temperature-sensitive mutants with defects in the lytic pathway. Virology. 1980 Aug;105(1):52–59. doi: 10.1016/0042-6822(80)90155-5. [DOI] [PubMed] [Google Scholar]
- Scott J. R. A turbid plaque-forming mutant of phage P1 that cannot lysogenize Escherichia coli. Virology. 1974 Dec;62(2):344–349. doi: 10.1016/0042-6822(74)90397-3. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Clear plaque mutants of phage P1. Virology. 1970 May;41(1):66–71. doi: 10.1016/0042-6822(70)90054-1. [DOI] [PubMed] [Google Scholar]
- Shepard H. M., Polisky B. Measurement of Plasmid copy number. Methods Enzymol. 1979;68:503–513. doi: 10.1016/0076-6879(79)68039-4. [DOI] [PubMed] [Google Scholar]
- Som T., Sternberg N., Austin S. A nonsense mutation in bacteriophage P1 eliminates the synthesis of a protein required for normal plasmid maintenance. Plasmid. 1981 Mar;5(2):150–160. doi: 10.1016/0147-619x(81)90016-0. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Powers M., Yarmolinsky M., Austin S. Group Y incompatibility and copy control of P1 prophage. Plasmid. 1981 Mar;5(2):138–149. doi: 10.1016/0147-619x(81)90015-9. [DOI] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid. 1977 Nov;1(1):1–7. doi: 10.1016/0147-619x(77)90003-8. [DOI] [PubMed] [Google Scholar]
- Vapnek D., Rupp W. D. Asymmetric segregation of the complementary sex-factor DNA strands during conjugation in Escherichia coli. J Mol Biol. 1970 Nov 14;53(3):287–303. doi: 10.1016/0022-2836(70)90066-5. [DOI] [PubMed] [Google Scholar]
- Walker D. H., Jr, Walker J. T. Genetic studies of coliphage P1. III. Extended genetic map. J Virol. 1976 Oct;20(1):177–187. doi: 10.1128/jvi.20.1.177-187.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun T., Vapnek D. Electron microscopic analysis of bacteriophages P1, P1Cm, and P7. Determination of genome sizes, sequence homology, and location of antibiotic-resistance determinants. Virology. 1977 Mar;77(1):376–385. doi: 10.1016/0042-6822(77)90434-2. [DOI] [PubMed] [Google Scholar]



