Abstract
Nonsteroidal anti-inflammatory drugs (NSAIDs) are well-known cancer preventives, which have been largely attributed to their antiproliferative and apoptosis-inducing activities. In this study, we show that microsatellite instability (MSI) in colorectal cancer cells deficient for a subset of the human mismatch repair (MMR) genes (hMLH1, hMSH2, and hMSH6), is markedly reduced during exposure to aspirin or sulindac [or Clinoril, which is chemically related to indomethacin (Indocin)]. This effect was reversible, time and concentration dependent, and appeared independent of proliferation rate and cyclooxygenase function. In contrast, the MSI phenotype of a hPMS2-deficient endometrial cancer cell line was unaffected by aspirin/sulindac. We show that the MSI reduction in the susceptible MMR-deficient cells was confined to nonapoptotic cells, whereas apoptotic cells remained unstable and were eliminated from the growing population. These results suggest that aspirin/sulindac induces a genetic selection for microsatellite stability in a subset of MMR-deficient cells and may provide an effective prophylactic therapy for hereditary nonpolyposis colorectal cancer kindreds where alteration of the hMSH2 and hMLH1 genes are associated with the majority of cancer susceptibility cases.
Hereditary nonpolyposis colorectal cancer (HNPCC) is one of the most frequent cancer predisposition syndromes and is characterized by early onset of colorectal cancers as well as cancers of the endometrium, stomach, and upper urinary tract (1–3). Germ-line mutations in two genes, hMSH2 and hMLH1, account for the vast majority of HNPCC cases, whereas mutations in the hPMS2 and hMSH6 genes are rare (4–8). These genes encode homologs of the prototypical bacterial MutS and MutL mismatch repair (MMR) proteins; at present there are upwards of 40 relatives in a variety of organisms, which have been identified as MutS homologs (MSH) or MutL homologs [MLH or postmeiotic segregant (PMS)] (9–11). The Saccharomyces cerevisiae and human counterparts have been implicated in nuclear and organellar MMR as well as distinctive meiotic activities (9–11).
In bacteria, yeast, and most of the HNPCC-related colorectal adenocarcinomas, instability of simple repeat sequences [microsatellite instability (MSI)] is an indicator of the loss of MMR function (12–14). Furthermore, elevated spontaneous mutation rates at all gene loci examined have been found to thoroughly correlate with the MSI phenotype (15, 16), which has lent support to the mutator hypothesis of tumorigenesis (17). Although MSI in proliferating cell lines was thought to be independent of doubling time, it recently has been shown that the mutator phenotype of two hMSH2-deficient cell lines was even more pronounced if cells were grown under confluent reduced growth conditions (18). Thus, MSI also may be a conditional event that depends on the growth state.
Recently, we devised a reliable diagnostic methodology for examining MSI in human tumors (19). This technology has led to the subsequent development of a rapid detection methodology for MSI in cell lines that is based on a modified primer-extension preamplification (PEP)-PCR technology (20). These techniques have allowed us to accurately measure the MSI frequency in five human cell lines containing defined mutations in MMR genes (21, 22): HCT116 (hMLH1−), LoVo (hMSH2−), HCT15 (hMSH6−), and HEC-1-A (hPMS2−). SW480, which is wild type for MMR, served as a control. In this study we show that treatment of colon tumor cell lines containing defects in hMLH1, hMSH2, and hMSH6, by aspirin or sulindac, dramatically reduces the MSI phenotype that typifies MMR defects. The effect was time and dose dependent and independent of proliferation and cyclooxygenase (COX) function. In contrast, similar treatment of the endometrial tumor cell line HEC-1-A (hPMS2−), had little or no effect on MSI. We show that the induction of apoptosis by aspirin/sulindac in the susceptible cell lines incites a genetic selection for microsatellite stability.
METHODS
Cell Lines and Cell Culture.
The human colon cancer cell lines SW480, LoVo, HCT116, and HCT15 were obtained from the American Type Culture Collection. SW48 was purchased from the European Collection of Animal Cell Cultures (Salisbury, Wiltshire, U.K.). HEC-1-A was kindly provided by M. Thiel, Institute of Anatomy, University of Essen, Germany. HCT116 contains a base substitution that results in a termination signal at codon 252 (TCA → TAA), and SW48 lacks any known mutations in its coding sequence but does not appear to express hMLH1 as a result of cytosine methylation in the promotor region. LoVo contains a deletion of exon 4–6 of hMSH2. HCT15/DLD1 contains a 1-bp deletion at codon 222 of hMSH6, resulting in a nonsense mutation as well as a frameshift mutation at codon 1103, which causes a new stop-codon 9 bp downstream. HEC-1-A contains a truncation mutation of hPMS2 at codon 802. HCT 116 was maintained in DMEM/Ham′s F12, LoVo was maintained in Ham’s F-12 and SW480, and HCT15 were maintained in RPMI medium 1640 (Biochrom, Berlin), all media containing 10% fetal bovine serum (Sigma). SW48 was kept in Leibowitz medium with 10% fetal calf serum (FCS), and HEC-1-A was grown in McCoy’s 5A supplemented with 10% FCS.
Large-Scale Subcloning and Microclone Subcloning.
Large-scale subclones of the various cell lines were generated by seeding cells in a 96-multiwell plate at ≈1 cell/well. At that time the cell lines were in culture for approximately 40 generations. Fifteen to 20 colonies from each cell line were expanded, and chromosomal DNA was isolated and subjected to PCR. For microclone subcloning, cells were seeded and grown on coverslips for 3 days. Distinct clones comprising groups of about 3–10 cells were picked under a microscope by using a sterile canula (microlance3, Becton Dickinson) and then lysed in 10 μl of digestion buffer (1× Expand HiFi PCR buffer no. 3, 4 mg/ml of proteinase K, and 5% Tween-20) for 16 hr at 50°C, and cellular debris was pelleted by microcentrifugation.
MSI Analysis.
The entire microclone-derived genomic DNA was subjected to random DNA amplification by using a modified PEP-PCR protocol according to Zhang et al. (20) (W.D., A. Hartmann, S.W., E. Heinmöller, T. Kerner, E. Endl, K.-W. Jauch, F.H. and J.R., unpublished work). Briefly, lysed clarified samples were added to 50 μl of PEP mix [0.05 mg/ml gelatine, 16 μM 15-mer random-primer, 0.1 mM dNTP, 3.6 units of Expand high-fidelity polymerase (Boehringer Mannheim), 2.5 mM MgCl2, in 1× Expand HiFi PCR buffer no. 3]. Amplification was performed for 50 cycles, each consisting of 94°C, 1 min; 37°C, 2 min; ramping of 0.1°C/sec to 55°C; 55°C, 4 min, and 68°C, 30 sec.
MSI was assessed in 15–20 subclones (microclones) per cell line by using either 3 μl of DNA of the random PEP-PCR aliquots derived from the microcell subclones or 100 ng DNA from large-scale subclones, at different microsatellite loci (19): BAT25, BAT26 and BAT40 containing mononucleotides; D13S175, adenomatous polyposis coli (APC) (D5S346), Mfd28 (D10S89), D3S1283, Mfd15 (D17S250), D2S123, and D18S58, which are dinucleotide repeats; TATA box-binding protein (TBP) as a trinucleotide repeat as well as Mycl1 and retinoblastoma, which are tetranucleotide repeats. PCR amplifications were performed in a final volume of 20 μl in an MJ Research Thermocycler (PTC100, MJ Research, Watertown, MA) for 50 cycles: 94°C, 1 min; 50–60°C/1 min; 72°C, 1 min followed by a final extension at 72°C, 8 min. Subsequently 6 μl of PCR product was added to 2 μl of loading buffer (96% formamide, 1% Xylene Cyanol FF, 1% bromophenol blue, 10 mM EDTA, pH 8.0) denatured at 95°C and separated by gel electrophoresis using 6.7% polyacrylamide/50% urea gels (1 hr, 1,500 V, 50°C). Bands were detected by silver nitrate staining as described (19). Alternatively, automated microsatellite analysis was performed on an ABI373 stretch (Applied Biosystems) using gene scan (Applied Biosystems) software. These two detection techniques produced identical results. MSI was defined as the fraction of subclones from each cell line exhibiting novel alleles at the different loci. Differences between the MSI frequency in treated and control lines were evaluated by Wilcoxon test for statistical significance (level was set to P < 0.05).
Nonsteroidal Anti-Inflammatory Drug (NSAID) Treatment.
Sulindac and aspirin (Sigma) were dissolved in 1 M Tris⋅HCl, pH 7.5 to a stock concentration of 100 mM and 1 M, respectively, and the pH was adjusted to 7.2 with 4 M HCl. For cellular treatment, 40 μl of 100 mM sulindac or 25 μl of 1M aspirin was added to 10 ml of culture medium, resulting in a final concentration of 400 μM sulindac and 2.5 mM aspirin (the increase in total salt concentration of the cell culture medium by the addition of aspirin/sulindac was negligible). Control cells were treated with equivalent quantity of 1 M Tris⋅HCl, pH 7.2 buffer without NSAIDs. At these concentrations of sulindac and aspirin the viability of the tumor cell lines tested was greater than 50% after 2 days of treatment, as assessed by means of the 3-(4,5-dimethylthiazol2-yl)-2,5 diphenyltetrazoliumbromid assay (for details see ref. 47). In the experiments shown in Figs. 2–5, cells were treated for approximately 3 months with 400 μM sulindac or 2.5 mM aspirin, which corresponds to about 90 total cell divisions. The time dependency of NSAIDs on MSI frequency was determined by using the microclone technique at 0, 5, 7, 14, 28, 56, 70, 84, and 112 days after continuous sulindac or aspirin treatment. The doubling time for the HCT116 (hMLH1−) cell line at the highest doses of both aspirin or sulindac was reduced from approximately 20 hr (untreated) to approximately 30.5 hr (treated). Thus, at 16 weeks, the untreated cells had progressed through approximately 130 generations, whereas the treated cells had progressed through approximately 90 generations. Serum starvation of untreated HCT116 (hMLH1−) cells resulted in an approximately 30-hr generation time. For demonstration of dose response, cells were treated for 12 weeks by using different concentrations of sulindac (40, 400, and 600 μM) or aspirin (1 and 2.5 mM), respectively.
Figure 2.
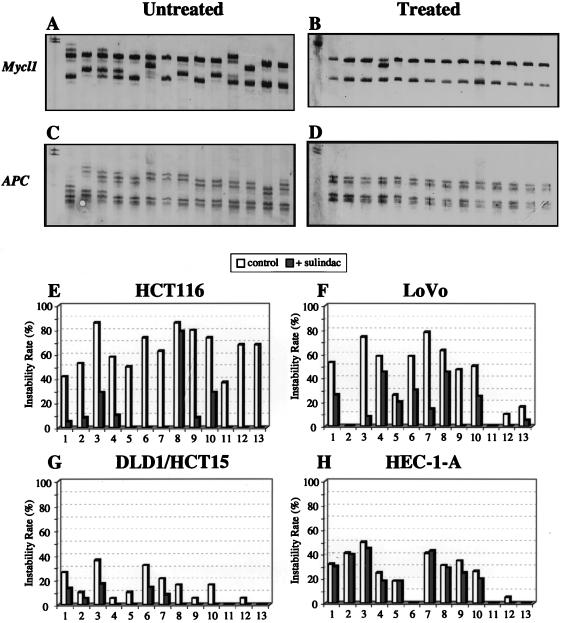
Effect of long-term sulindac treatment on MSI frequency. MSI frequency in HCT116 cells after 16 weeks of 400 μM sulindac treatment comparing Mycl1 locus (A, untreated; B, treated) and APC locus (C, untreated; D, treated). Thirteen microsatellite loci (1, BAT25; 2, BAT26; 3, BAT40; 4, D13S175; 5, APC/D5S346; 6, Mfd28/D10S89; 7, D3S1287; 8, Mfd15/D17S250; 9, D2S123; 10, D18S58; 11, TATA box-binding protein; 12, Mycl1; 13, Rb) and three cell lines were examined after treatment with sulindac (400 μM). (E) HCT116 (hMLH1−). (F) Lovo (hMSH2−). (G) HCT15/DLD1 (hMSH6−). (H) HEC-1-A (hPMS2−). The primary panel of five diagnostic microsatellite loci recommended by the NCI/ICG-HNPCC (20) corresponds to numbers 1, 2, 5, 8, and 9. In E-H the empty bars indicate MSI in untreated controls and filled bars indicate MSI after treatment with sulindac.
Figure 5.
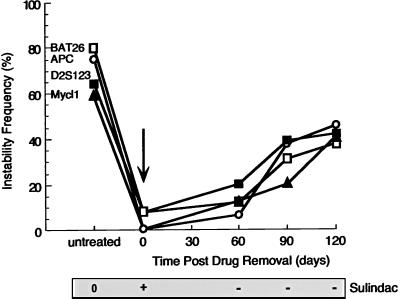
Kinetics analysis of the MSI phenotype recovery after withdrawal of sulindac. HCT116 cells were treated with sulindac (400 μM) for 16 weeks and then at time 0 sulindac was withdrawn. MSI at three loci (BAT26, APC, and Mycl1) was followed for an additional 3.5 months.
Apoptosis Determination.
The kinetics of apoptosis after continuous sulindac treatment or serum starvation were studied in HCT116 (hMLH1−) cells 2, 5, 14, 28, 70, and 112 days by using the Cell Death Detection ELISAPLUS Kit (Boehringer Mannheim). ELISA OD readings were correlated with the number of apoptotic cells by terminal deoxynucleotidyltransferase-mediated UTP end labeling (TUNEL) assay (2.5 OD ≈30% apoptotic cells). The Annexin Kit (Boehringer Mannheim) was used as a primary selection system for apoptotic cell separation via FACS. Briefly, HCT116 cells were grown in the presence of 400 μM sulindac, split, and regrown approximately 12 times over the course of a 12-week treatment. After treatment, the cells were stained with the annexin primary antibody and counterstained with a fluorescent probe, and annexin-positive (apoptotic) cells were separated by FACS into eight separate five-cell pools. An analogous set of five-cell pools was obtained from untreated controls (nonapoptotic), and both were subjected to genomic DNA isolation and PEP-PCR as described. These results were independently confirmed by using the TUNEL assay (POD, Boehringer Mannheim). For TUNEL identification, 5 × 103 HCT116 (hMLH1−) cells were exponentially grown on chamber slides for 48 hr. Apoptotic cells were identifiable by brown nuclear TUNEL staining. These cells as well as nonapoptotic control cells then were individually picked and analyzed for MSI at BAT26, Mycl1, and APC locus before treatment and after 2 weeks and 12 weeks of sulindac treatment.
RESULTS
MSI in Tumor Cell Lines.
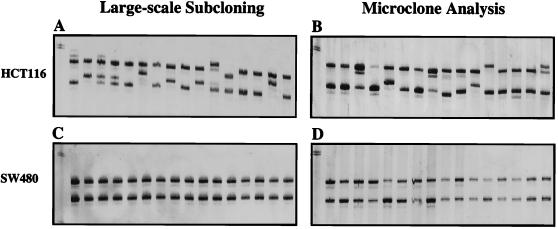
The efficacy of our MSI technology was evaluated in cell lines by comparing traditional large-scale clonal outgrowths derived from a single or small number of cells to a more rapid method, which examines small isolated microclones of cells grown on coverslips that then are individually picked under a microscope. We found that both methods produced identical results (Fig. 1). Examination of 13 microsatellite loci, which included the primary panel of five National Cancer Institute (NCI)/International Collaborative Group for HNPCC (ICG-HNPCC)‖-designated MSI diagnostic loci (19), supports previous reports that showed that the frequency of MSI depended on the type of MMR gene mutation (Fig. 2 E-G, untreated controls). The hMLH1-deficient HCT-116 cell line displayed the highest MSI frequency (61%), followed by the hMSH2-deficient cell line LoVo (44%), then the hPMS2-deficient cell line HEC-1-A (38%) and the hMSH6-deficient cell lines HCT15/DLD1 (14%), which displayed relatively low MSI. Overall, the frequency of mono- and dinucleotide repeat instability was higher than tri- or tetranucleotide repeat instability (Fig. 2 E-G, untreated controls). Furthermore, as would have been expected from previous reports, the DLD1/HCT15 (hMSH6−) cell line showed largely mononucleotide repeat instability, which artificially lowered its overall MSI frequency (Fig. 2 E-G, untreated controls). These results confirm our data in human colorectal carcinomas as well as similar data in yeast, where mononucleotides and dinucleotides with complex repeat motifs were hotspots of MSI (19, 23). The MMR-proficient cell line SW480 displayed no MSI (Fig. 1C). We also confirmed that the MMR-deficient cell lines HCT116 (hMLH1−), LoVo (hMSH2−), HCT15 (hMSH6−), and HEC-1-A (hPMS2−) have no detectable MMR protein of the correspondingly altered MMR gene by Western analysis (data not shown).
Figure 1.
Analysis of microclone assay for MSI. MSI in subclones of HCT116 (A and B) and SW480 (C and D). MSI analysis at Mycl1 locus in HCT116 cells performed by large-scale subcloning (A) and microclone subcloning (B). MSI analysis at the Mycl1 locus in wild-type SW480 control cell line performed by large-scale subcloning (C) and microclone subcloning (D).
Aspirin and Sulindac Affect MSI.
Because MSI evaluated by the microclone assay produced the same result as large-scale subcloning analysis, we used the more expeditious microclone technology to examine parameters that might influence the MSI mutator phenotype. NSAIDs have been shown to display cancer preventive and tumor regressive effects in colon cancer (24–28). Several epidemiological studies have shown that prolonged use of aspirin is associated with reduced risk of colorectal cancer by as much as 40–50% and also appears efficacious in other gastrointestinal cancers, such as esophageal and gastric carcinoma as well as several other tumor types (29–34). Most interestingly, it has been shown that NSAIDs, particularly sulindac, induce regression of adenomas in familial adenomatous polyposis patients (35).
Similarly, we observed a marked reduction of the MSI frequency in the HCT116 (hMLH1−), Lovo (hMSH2−), and DLD1/HCT15 (hMSH6−) colon tumor cell lines after treatment with either aspirin or sulindac (Fig. 2). This effect could be demonstrated in all five NCI/ICG-HNPCC diagnostic microsatellite loci and was most striking at the Mycl1 and BAT26 locus (Fig. 2 A-D) (19). The reduction of MSI frequency was more pronounced in the hMLH1-deficient cell lines than in the hMSH2- or hMSH6-deficient cell lines (Fig. 2 E-G). In contrast, the HEC-1-A hPMS2-deficient endometrial tumor cell line displayed little or no significant change of MSI frequency after aspirin/sulindac treatment (Fig. 2H). It is worth emphasizing that, unlike the colorectal cell lines used in the majority of this study, HEC-1-A (hPMS2−) was derived from an endometrial tumor.
Mechanism of MSI Suppression.
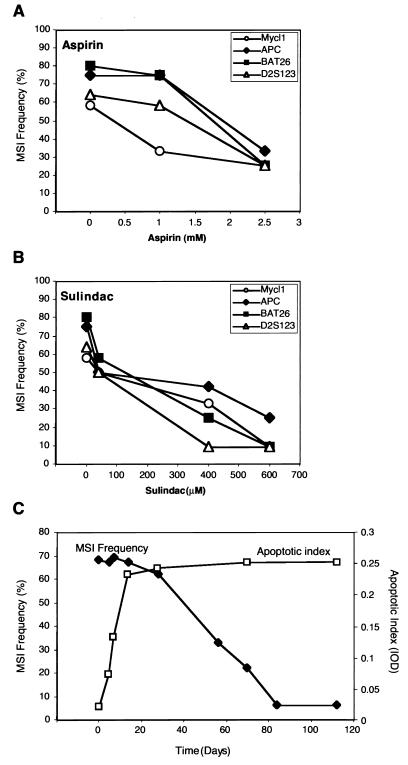
To investigate the mechanism of NSAIDs suppression of MSI, we have focused on the HCT116 (hMLH1−) cell line as a model because of its robust growth and potent response to aspirin/sulindac. The reduction in MSI frequency in the HCT116 (hMLH1−) cell line was found to be time and dose dependent (Fig. 3). No significant decrease of MSI frequency could be observed until 8 weeks of culture (Fig. 3C). However, after 12 and 16 weeks of culture a maximal suppression of the mutator phenotype was achieved, with the MSI frequency often 10 times lower than in the control untreated cells. In addition, suppression of the mutator phenotype appeared entirely dependent on the concentration of aspirin and sulindac (Fig. 3 A and B). We were unable to investigate a wide range of aspirin or sulindac concentrations, because drug-induced cell death exceeded 50% at doses above those shown. Examination of the unaltered MMR proteins in these cell lines suggested that their levels remain constant during the treatment period, eliminating expression compensation as a mechanism for this effect (data not shown).
Figure 3.
Time and concentration dependence of NSAID treatment on MSI frequency. (A) Concentration dependence of MSI reduction by aspirin. (B) Concentration dependence of MSI reduction by sulindac, in HCT116 (hMLH1−) cells after 12 weeks of treatment. (C) Time dependency of MSI frequency (filled diamond) and apoptotic index (squares) in HCT116 cells during treatment with sulindac (400 μM). Comparison of apoptotic index assay (derived from spectrophotometric ELISA readings) to TUNEL assay (where apoptotic cells are counted under a microscope) we estimate that an apoptotic index of 0.25 corresponds to approximately 30% apoptotic cells. For untreated cells the percentage of apoptotic cells is 1% or lower.
Both aspirin and sulindac have been reported to display profound antiproliferative effects on tumor cell lines, alter the cell cycle distribution, and induce apoptosis (36–38). By using flow cytometry we confirmed the well-known observation that cells treated with aspirin and sulindac accumulate at G0/G1 (data not shown). These results suggested that the reduction of MSI frequency might merely be a consequence of reduced proliferation rate. To address this possibility we determined the MSI frequency in untreated serum-starved HCT116 (hMLH1−) cells. Both the serum-starved and aspirin/sulindac-treated HCT116 (hMLH1−) cells showed a decrease in doubling time from approximately 20 hr to 30 hr with no apparent change in the length of S-phase as determined by BrdUrd incorporation (data not shown). We found no significant changes in the frequency of MSI when we compared these serum-starved HCT116 (hMLH1−) cells to normally grown HCT116 (hMLH1−) cells (data not shown). As a further control, we examined serum-starved as well as sulindac-treated (2 weeks) and untreated G0/G1 and S/G2/M HCT116 (hMLH1−) cells separated by FACS and also found no significant difference in MSI (data not shown). Although altered cellular proliferation induced by aspirin/sulindac treatment versus serum starvation might not be functionally equivalent, they appear quite similar by all of the parameters that we have examined. Thus, it is unlikely that the decrease in MSI frequency is the result of any antiproliferative effects induced by NSAID treatment. As a further support, Richards et al. (18) demonstrated an increase in MSI frequency when they examined subclones of hMSH2-deficient cell lines that were maintained at high density under greatly diminished proliferation conditions.
The best-known mechanism of NSAIDs is the inhibition of COX, the enzyme that converts arachidonic acid to prostanoids in the inflammatory response (26, 39). Two COX genes have been cloned (COX-1 and COX-2). COX-1 is ubiquitously expressed and appears to be a housekeeping gene, whereas COX-2 expression is inducible by growth factors and oncogenes (40). Interestingly, it has been shown that tumors contain high-level expression of COX-2, which appears to inhibit apoptosis in colon carcinogenesis (41, 42); the chemopreventive effect of NSAIDs has been largely attributed to this gene (26, 39). A direct link between COX genes and the MSI mutator phenotype is unlikely, because we observed a clear reduction of MSI frequency after aspirin and sulindac treatment in DLD1/HCT15 cells (Fig. 2C), which lacks expression of both of the known COX genes (43).
Aspirin/Sulindac-Induced Apoptosis Results in a Genetic Selection.
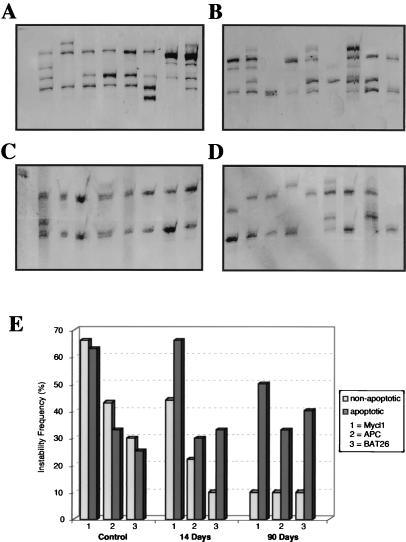
The induction of apoptosis has been proposed to contribute to the chemopreventive properties of NSAIDs (44, 45). Western blot and immunocytochemical analyses of the NSAID-treated cell population suggest that, whereas p53 status is largely unchanged, the p21 and BAX proteins are markedly induced (data not shown). We examined the onset of apoptosis directly and found that treatment of cells with aspirin and sulindac induced apoptosis very rapidly, reaching a maximum after 2 weeks of treatment (Fig. 3C). In contrast, we observed a decrease in the frequency of MSI in response to aspirin or sulindac treatment, no earlier than 8 weeks of treatment, with lowest MSI frequency after 12–16 weeks (Fig. 3C). As a first step in understanding the contribution of apoptosis to the reduction in MSI after NSAID treatment, we examined MSI at three NCI/ICG-HNPCC diagnostic microsatellite loci in apoptotic and nonapoptotic cells (Fig. 4 A-C). Both apoptotic and nonapoptoic cells were separated into five-cell pools via FACS by using the annexin staining system as a primary marker for the early events associated with apoptosis (Fig. 4 A and B). Examination of MSI by this methodology is equivalent to the limiting-dilution MSI technique described previously (46). Remarkably, MSI remained constant in the apoptotic cells, whereas MSI gradually decreased in the nonapoptotic cells (Fig. 4C). This finding should be contrasted to serum starvation where the number of apoptotic cells increases to nearly the same level as cells treated with NSAIDs, yet there is no difference in MSI between the apoptotic and nonapoptotic fractions (data not shown). These results suggest that treatment by aspirin/sulindac induces a genetic selection for cells that retain stable microsatellites. This observation was independently confirmed by single-cell isolation and PEP-PCR after TUNEL staining to determine apoptotic and nonapoptotic cells (data not shown). Furthermore, withdrawal of aspirin or sulindac from MMR-deficient cells resulted in a return of the MSI mutator phenotype with kinetics that were similar to the treatment of virgin cells with aspirin (Fig. 5). These results suggest that the MSI suppression is reversible, that constant exposure to aspirin/sulindac is required to maximize efficacy, and is consistent with the idea that aspirin/sulindac exposure promotes an obligate genetic selection for microsatellite stability.
Figure 4.
Genetic selection of MSI via apoptosis. Representative gel analysis of MSI alterations at the Mycl1 locus in untreated HCT116 (hMLH1−) nonapoptotic (A) and apoptotic cells (B) and HCT116 (hMLH1−) nonapoptotic (C) and apoptotic (D) cells after a 12-week treatment with sulindac (400 μM). (E) Graphic analysis of the MSI frequency alteration in HCT116 (hMLH1−) cells at the BAT26, APC, and Mycl1 loci during a 12-week sulindac treatment (400 μM).
DISCUSSION
Suppression of the MSI mutator phenotype by aspirin/sulindac is extraordinary and suggests a prophylactic treatment for the genetic susceptibility to cancer displayed by a subset of HNPCC families (particularly those with an inherited alteration of hMSH2, hMSH6, and hMLH1). The efficacy of this prophylactic treatment is likely to depend on whether aspirin affects the frequency of the “second hit” required to express the tumorigenic phenotype in heterozygous carriers of HNPCC alterations, as well as suppression of the mutator phenotype associated with dysfunction of the MMR genes in neoplasms. Evaluation of this prophylactic therapy in HNPCC carriers has not been performed. Our studies also appear to suggest that a component of the cancer-preventive effect observed in the general population may be the result of specific effects on tumorigenesis associated with alterations in the MMR genes. This antitumorigenic effect is likely to be different from the therapeutic effect observed in clinical trials with familial adenomatous polyposis patients where there is a dramatic decrease in adenomatous tissue after treatment with sulindac.
The pathway for genetic selection that results in MSI suppression after exposure to aspirin/sulindac in the colorectal tumor cell lines is unknown. It is possible that aspirin/sulindac alters the recognition of threshold genetic instability as well as the subsequent apoptotic decision in a subset of MMR-deficient cells or aspirin/sulindac may induce genetic stability by affecting the fidelity of the DNA metabolic machinery. Although the apoptotic decision through the Fas death-receptor pathway is well known, the apoptotic decision initiated by threshold DNA damage and/or instability remains obscure. We entertain the possibility that the MMR system is a sensor for threshold genomic instability leading to apoptosis. In this model, the effect of aspirin/sulindac would be to circumvent the absence of such a threshold-instability apoptotic signal, normally controlled by the MMR machinery, perhaps to a secondary pathway. The observation that MSI in hPMS2-deficient cells is not significantly suppressed by aspirin/sulindac may be relevant because our results would appear to suggest that functional hPMS2 is necessary for the genetic selection induced by exposure to aspirin/sulindac or that PMS2 is outside the MMR threshold-instability apoptotic pathway. It is important to note that the hPMS2-deficient HEC-1-A cell line is derived from an endometrial tumor that is inherently different from the hMLH1-, hMSH2-, and hMSH6-deficient colorectal tumor cell lines. Regardless, the genetic selection imposed by aspirin/sulindac would clearly affect the majority of HNPCC tumors, which can be largely accounted for by alterations of hMSH2 and hMLH1. Finally, whereas a dissociation of MSI and elevated global spontaneous mutation rates never have been observed, it is possible that the effects of aspirin/sulindac could be confined to the suppression of MSI whereas the widespread increase in spontaneous mutation rates characteristic of MMR defects may remain unperturbed. Tests of these hypotheses are underway.
Acknowledgments
We are grateful to Christoph Schmutte for helpful discussions and assistance with figures and Hansjorg Alder and the Kimmel Nucleic Acids Facility for automated microsatellite analysis. T.B. was supported by Grant Bo/1445–2 from the Deutsche Forschungsgemeinschaft (DFG). This work was supported in part by Wilhelm Sander-Stiftung (93.055.2), Munich, Germany (J.R.) and National Institutes of Health Grants CA56542 and CA67007 (R.F.).
ABBREVIATIONS
- NSAID
nonsteroidal anti-inflammatory drug
- MSI
microsatellite instability
- MMR
mismatch repair
- HNPCC
hereditary nonpolyposis colorectal cancer
- MSH
MutS homolog
- MLH
MutL homolog
- PMS
postmeiotic segregant
- PEP
primer-extension preamplification
- APC
adenomatous polyposis coli
- TUNEL
terminal deoxynucleotidyltransferase-mediated UTP end labeling
- NCI
National Cancer Institute
- ICG-HNPCC
International Collaborative Group for HNPCC
- COX
cyclooxygenase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Definition of microsatellite panel and MSI diagnostic definition was determined at a December 8–9, 1998 meeting sponsored by the NCI/ICG-HNPCC.
References
- 1.Dunlop M G, Farrington S M, Carothers A D, Wyllie A H, Sharp L, Burn J, Liu B, Kinzler K W, Vogelstein B. Hum Mol Genet. 1997;6:105–110. doi: 10.1093/hmg/6.1.105. [DOI] [PubMed] [Google Scholar]
- 2.Hodgson S V, Bishop D T, Dunlop M G, Evans D G, Northover J M. J Med Screen. 1995;2:45–51. doi: 10.1177/096914139500200112. [DOI] [PubMed] [Google Scholar]
- 3.Lynch H T, Smyrk T, Lynch J. Cancer Genet Cytogenet. 1997;93:84–99. doi: 10.1016/s0165-4608(96)00290-7. [DOI] [PubMed] [Google Scholar]
- 4.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 5.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A, et al. Nature (London) 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 6.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, et al. Nature (London) 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 7.Miyaki M, Konishi M, Tanaka K, Kikuchi Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, Yuasa Y. Cancer Res. 1997;57:3920–3923. [PubMed] [Google Scholar]
- 9.Fishel R, Kolodner R D. Curr Opin Genet Dev. 1995;5:382–395. doi: 10.1016/0959-437x(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 10.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 11.Fishel R, Wilson T. Curr Opin Genet Dev. 1997;7:105–113. doi: 10.1016/s0959-437x(97)80117-7. [DOI] [PubMed] [Google Scholar]
- 12.Ionov Y, Peinado M A, Malkbosyan S, Shibata D, Perucho M. Nature (London) 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 13.Thibodeau S N, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 14.Aaltonen L A, Peltomaki P, Leach F, Sistonen P, Pylkkanen S M, Mecklin J-P, Jarvinen H, Powell S, Jen J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharyya N P, Skandalis A, Ganesh A, Groden J, Meuth M. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eshleman J R, Markowitz S D, Donover P S, Lang E Z, Lutterbaugh J D, Li G M, Longley M, Modrich P, Veigl M L, Sedwick W D. Oncogene. 1996;12:1425–1432. [PubMed] [Google Scholar]
- 17.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 18.Richards B, Zhang H, Phear G, Meuth M. Science. 1997;277:1523–1526. doi: 10.1126/science.277.5331.1523. [DOI] [PubMed] [Google Scholar]
- 19.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 20.Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyer J C, Umar A, Risinger J I, Lipford J R, Kane M, Yin S, Barrett J C, Kolodner R D, Kunkel T A. Cancer Res. 1995;55:6063–6070. [PubMed] [Google Scholar]
- 22.Glaab W E, Tindall K R. Carcinogenesis. 1997;18:1–8. doi: 10.1093/carcin/18.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Sia E A, Kokoska R J, Dominska M, Greenwell P, Petes T D. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henschke U K, Luande G J, Choppala J D. J Natl Med Assoc. 1977;69:581–584. [PMC free article] [PubMed] [Google Scholar]
- 25.Baron J A. Preventive Med. 1995;24:121–124. doi: 10.1006/pmed.1995.1023. [DOI] [PubMed] [Google Scholar]
- 26.DuBois R N, Smalley W E. J Gastroenterol. 1996;31:898–906. doi: 10.1007/BF02358623. [DOI] [PubMed] [Google Scholar]
- 27.Thun M J. Gastroenterol Clin N Amer. 1996;25:333–348. doi: 10.1016/s0889-8553(05)70250-8. [DOI] [PubMed] [Google Scholar]
- 28.Williams C S, Smalley W, DuBois R N. J Clin Invest. 1997;100:1325–1329. doi: 10.1172/JCI119651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thun M J, Namboodiri M M, Calle E E, Flanders W D, Heath C W., Jr Cancer Res. 1993;53:1322–1327. [PubMed] [Google Scholar]
- 30.Suh O, Mettlin C, Petrelli N J. Cancer. 1993;72:1171–1177. doi: 10.1002/1097-0142(19930815)72:4<1171::aid-cncr2820720407>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 31.Schreinemachers D M, Everson R B. Epidemiology. 1994;5:138–146. doi: 10.1097/00001648-199403000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Funkhouser E M, Sharp G B. Cancer. 1995;76:1116–1119. doi: 10.1002/1097-0142(19951001)76:7<1116::aid-cncr2820760703>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 33.Egan K M, Stampfer M J, Giovannucci E, Rosner B A, Colditz G A. J Natl Cancer Inst. 1996;88:988–993. doi: 10.1093/jnci/88.14.988. [DOI] [PubMed] [Google Scholar]
- 34.Cramer D W, Harlow B L, Titus Ernstoff L, Bohlke K, Welch W R, Greenberg E R. Lancet. 1998;351:104–107. doi: 10.1016/S0140-6736(97)08064-1. [DOI] [PubMed] [Google Scholar]
- 35.Giovannucci E, Rimm E B, Stampfer M J, Colditz G A, Ascherio A, Willett W C. Ann Intern Med. 1994;121:241–246. doi: 10.7326/0003-4819-121-4-199408150-00001. [DOI] [PubMed] [Google Scholar]
- 36.Shiff S J, Qiao L, Tsai L L, Rigas B. J Clin Invest. 1995;96:491–503. doi: 10.1172/JCI118060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricchi P, Pignata S, Di Popolo A, Memoli A, Apicella A, Zarrilli R, Acquaviva A M. Int J Cancer. 1997;73:880–884. doi: 10.1002/(sici)1097-0215(19971210)73:6<880::aid-ijc20>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 38.Sheng H, Shao J, Kirkland S C, Isakson P, Coffey R J, Morrow J, Beauchamp R D, DuBois R N. J Clin Invest. 1997;99:2254–2259. doi: 10.1172/JCI119400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gustafson Svard C, Lilja I, Hallbook O, Sjodahl R. Ann Med. 1997;29:247–252. doi: 10.3109/07853899708999342. [DOI] [PubMed] [Google Scholar]
- 40.Prescott S M, White R L. Cell. 1996;87:783–786. doi: 10.1016/s0092-8674(00)81983-2. [DOI] [PubMed] [Google Scholar]
- 41.Tsujii M, DuBois R N. Cell. 1995;83:493–501. doi: 10.1016/0092-8674(95)90127-2. [DOI] [PubMed] [Google Scholar]
- 42.Sano H, Kawahito Y, Wilder R L, Hashiramoto A, Mukai S, Asai K, Kimura S, Kato H, Kondo M, Hla T. Cancer Res. 1995;55:3785–3789. [PubMed] [Google Scholar]
- 43.Hanif R, Pittas A, Feng Y, Koutsos M I, Qiao L, Staiano Coico L, Shiff S I, Rigas B. Biochem Pharmacol. 1996;52:237–245. doi: 10.1016/0006-2952(96)00181-5. [DOI] [PubMed] [Google Scholar]
- 44.Shiff S J, Koutsos M I, Qiao L, Rigas B. Exp Cell Res. 1996;222:179–188. doi: 10.1006/excr.1996.0023. [DOI] [PubMed] [Google Scholar]
- 45.Qiao L, Hanif R, Sphicas E, Shiff S J, Rigas B. Biochem Pharmacol. 1998;55:53–46. doi: 10.1016/s0006-2952(97)00400-0. [DOI] [PubMed] [Google Scholar]
- 46.Parsons R, Li G-M, Longley M, Modrich P, Liu B, Berk T, Hamilton S R, Kinzler K W, Vogelstein B. Science. 1995;268:738–740. doi: 10.1126/science.7632227. [DOI] [PubMed] [Google Scholar]
- 47.Mosmann T. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]