Abstract
Mice lacking desmin produce muscle fibers with Z disks and normal sarcomeric organization. However, the muscles are mechanically fragile and degenerate upon repeated contractions. We report here a human patient with severe generalized myopathy and aberrant intrasarcoplasmic accumulation of desmin intermediate filaments. Muscle tissue from this patient lacks the wild-type desmin allele and has a desmin gene mutation encoding a 7-aa deletion within the coiled-coil segment of the protein. We show that recombinant desmin harboring this deletion cannot form proper desmin intermediate filament networks in cultured cells, nor is it able to assemble into 10-nm filaments in vitro. These findings provide direct evidence that a mutation in desmin can cause human myopathies.
A member of the intermediate filament (IF) superfamily, desmin forms 10-nm filaments in skeletal, cardiac, and smooth muscle cells. It is expressed earlier in embryogenesis than many other muscle-specific proteins, including myosins and α-actin (1). In maturing skeletal and cardiac muscle cells, desmin IFs organize at the peripheries of the aligning Z disks that anchor the barbed ends of actin thin filaments to the ends of each sarcomere (2, 3). By virtue of their location, desmin IFs have been implicated in laterally organizing the striated myofibrils and in linking sarcomeres to the sarcolemma membrane (4).
IFs are thought to act as mechanical integrators of cytoplasm, a feature that is particularly important in tissues subjected to repeated mechanical stress (for review, see ref. 5). Thus, for example, mice lacking basal epidermal IFs or expressing dominant negative-acting mutant basal epidermal keratin genes undergo skin blistering upon mild mechanical trauma (6, 7). These mice die within a few days after birth (7). In humans, this cell degenerative condition is known as epidermolysis bullosa simplex (EBS), and most EBS patients have mutations in one of their basal epidermal keratin genes (8–10). Many other genetic disorders of keratin have been elucidated, and these diseases similarly affect the mechanical integrity of cells (5). In these disorders, a marked correlation exists between the severity in phenotype, the location of the keratin mutation, and the degree to which the mutation affects IF assembly (5, 11).
Recent knockout studies in mice support the notion that desmin, like many keratins, plays a role in imparting mechanical stability to cells (12, 13). Desmin null mice are viable and fertile; however, all three muscle types develop regional signs of muscle weakness and degeneration accompanied by misaligned and/or split Z disks (13, 14). Defects are most prominent in highly contracting muscles, and physical measurements suggest that desmin functions in cellular transmission of both active and passive force (14, 15).
Desminopathies are a heterogeneous group of human myopathies characterized by abnormal aggregations of desmin in skeletal and/or cardiac muscle fibers (for review, see ref. 16). The clumping of desmin in muscle cells of these patients bears a striking resemblance to the keratin aggregates that accumulate in degenerating epithelial cells of various human keratin disorders (for review, see ref. 5). Although analysis of DNAs from three families with autosomal dominant desminopathy failed to show linkage to the desmin gene (17), the manifestations of desminopathies are variable, and the etiology remains unknown.
We have studied an unusual case of a desminopathy displaying skeletal, cardiac, and smooth muscle defects. The family had a history of cerebrovascular attack, and at 19 years, the patient exhibited generalized muscle weakness; at 28 years, respiratory failure and intestinal pseudoobstruction led to death (18).
We report here a homozygous deletion of 21 nt in the desmin gene(s) of this patient’s muscle. The mutation predicts a mutant desmin protein, lacking residues R173–E179. We provide functional analysis to demonstrate that this mutation severely compromises desmin’s ability to assemble into IFs in vivo and in vitro.
MATERIALS AND METHODS
Patient and Ultrastructural Analysis of Muscle Biopsies.
The clinical and pathologic features of the case have been detailed elsewhere (18). The patient’s family members and normal control subjects also were studied in this report. A right deltoid muscle biopsy was taken for diagnostic purposes with the informed consent of the patient. Immunohistochemistry revealed a patchy distribution of desmin (18). A portion of tissue was fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer, postfixed in 1.5% osmium tetroxide, and embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate and examined with a JEOL 1010 transmission electron microscope at 80 kv. Biopsies initially were examined by Ariza et al. (18) and reevaluated for the present study.
Isolation of Genomic DNAs and Desmin Exon Amplification.
Genomic DNAs were isolated from a skeletal muscle biopsy of the patient and from peripheral blood samples of family members and normal individuals. These DNAs served as templates for PCR amplification of exons and flanking intron segments of the desmin gene. The primer sets used were 3F/3R′ (first half of exon 1), 4F/4R (second part of exon 1), 5F/5R (exons 2–3), 6F/6R (exon 4), 7F/7R (exon 5), 8F/8R (exon 6), 9F/9R (exon 7), and 10F/10R (exons 8–9). All have been described (17) except 3R′: 5′-GCTGGACTTCTCACTGGCCG-3′. PCR cycling conditions were: 40 cycles of 95°C, 45 sec and 55°C, 1 min for 3FR, 6FR, and 8FR, and 40 cycles of 95°C, 45 sec and 60°C, 1 min for 4FR, 5FR, 7FR, 9FR, and 10 FR, using Taq polymerase Eurobiotaq (Eurobio, Paris).
Mutation Analysis of the Desmin Gene.
Amplified products were used for single-strand conformation polymorphism (SSCP) studies. For SSCP analysis, 2 μl of the PCR products were mixed with 6 μl of a 95% formamide, 0.05% xylene cyanol, 0.05% bromophenol blue, 20 mM EDTA solution, denatured by incubation at 80°C for 3 min and placed on ice. Electrophoresis was performed in a 10 × 7.5 cm nondenaturing polyacrylamide gel, run at 150 V for 2 h. Each sample was analyzed by electrophoresis under each of the following conditions: 8% or 12% acrylamide gels, with or without 10% glycerol, and at room temperature or 4°C. Gels were silver-stained (19) to visualize the bands.
PCR products exhibiting an aberrant SSCP shift were cloned into pUC18 by using a SureClone Ligation Kit (Pharmacia Biotech), and sequenced with an AbiPrism DNA Sequencing Kit (Perkin–Elmer). After identification of the patient mutation, exon 1 was amplified from DNAs of family members and 100 wild-type alleles. Samples then were run in a 6% acrylamide, 8 M urea denaturing gel and silver-stained.
Engineering of Expression Vectors.
Full-length hamster desmin cDNA (pUC19Des) was a gift of Hans Bloemendal (Univ. of Nijmegen, The Netherlands) (20, 21). Site-directed mutagenesis was used to introduce the 21-bp deletion mutation. A PCR product spanning nucleotides 474–731, but missing nucleotides 513–533 was cloned into the unique NarI and BsmI sites to create plasmid pUC19Δ7Des. All PCR products and engineered mutations were sequenced to confirm their identity.
The cDNAs in plasmids pUC19Des and pUC19Δ7Des were excised and cloned into cytomegalovirus (CMV) promoter expression vector pCB6+ to generate pCMVDes and pCMVΔ7Des. Plasmid pJVim, containing the human vimentin cDNA under the control of the simian virus 40 major early promoter (22) was used for expression of vimentin. Plasmids were purified twice by using CsCl density gradient ultracentrifugation.
For bacterial expression of proteins, the pET vector was used (23). An NdeI site was engineered at the start codon of pUC19Des cDNA by site-directed mutagenesis. This change did not alter the encoded amino acid sequence. A silent NotI site also was engineered at the desmin stop codon. The entire coding segment was ligated into the NdeI and NotI sites of pET23b+ (Novagen) to create pETDes. The NarI/BsmI fragment from pUC19Δ7Des then was cloned into pETDes to create pETΔ7Des. Proteins were purified by passaging the solubilized inclusion body fraction through an 8-ml Mono-Q anion exchange column, by using a buffer of 6 M urea, 50 mM Tris, 2 mM DTT, 0.3 mg/ml phenylmethylsulfonyl fluoride, 1 mM EGTA, pH 8.1 (24). Desmin was eluted with an 80-ml gradient of 0–175 mM guanidine HCl, run at 1 ml/min. Fractions were collected and kept at −80°C until needed.
Transient Transfection of Cultured Cells.
Human breast epithelial cells (MCF-7) and monkey COS epithelial cells were grown to 50% confluence and transfected by using Fugene 6 transfection medium (Boehringer Mannheim). At 48 h posttransfection, cells were fixed and examined by indirect immunofluorescence, by using (i) a mouse monoclonal anti-desmin antibody (Accurate Scientific, Westbury, NY), or (ii) a rabbit polyclonal anti-desmin (ICN) and a mouse monoclonal anti-vimentin antibody (Boehringer Mannheim). Secondary antibodies were either goat anti-mouse IgG or goat anti-rabbit IgG (Alexa, Molecular Probes).
In Vitro Filament Assembly.
Assembly of desmin filaments from purified recombinant protein was performed as described (25). Protein concentration was adjusted to 250 μg/ml, and aliquots were dialyzed against: (i) 8 M urea 5 mM Tris⋅HC1, 5 mM β-ME, pH 8.5, for 5 h at room temperature; (ii) 5 mM Tris⋅HCl, 5 mM β-mercaptoethanol, pH 8.5, for 12–16 h at 4°C; and (iii) 25 mM Tris⋅HCl, 160 mM NaCI, 5 mM β-ME, pH 7.5., for 5 h at room temperature. Filaments were examined by negative-staining (1% aqueous uranyl acetate). Polymerization efficiency was determined by subjecting final assemblies (40-μl aliquots, ≈8 μg proteins) to centrifugation at 100,000 × g for 60 min at 4°C (Airfuge, Beckman). Supernatant and pellet fractions were analyzed by SDS/PAGE and Coomassie blue staining.
Immunoblot Analysis.
Proteins were resolved by electrophoresis through 8.5% polyacrylamide denaturing gels, and gels were transferred by electroblotting to Immobilon P membranes (Millipore). Primary and secondary antibodies were a rabbit polyclonal antidesmin antibody (ICN) and a horseradish peroxidase-conjugated goat anti-rabbit IgG antibody, respectively. Blots were visualized by chemiluminescence (Amersham).
RESULTS
Muscle and Z-Band Abnormalities of a Patient With a Severe Generalized Myopathy.
Previous examination of this patient’s skeletal muscle, myocardium, and intestinal muscle revealed numerous atrophic fibers and frequent internalization of nuclei with subsarcolemmal eosinophilic masses (18). Immunohistochemistry was positive for desmin and negative for vimentin; antidesmin staining was patchy with extensive areas lacking staining and immunoreactive aggregates in other regions (18).
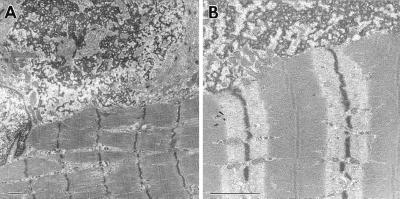
Ultrastructural analysis of postmortem skeletal muscle revealed sarcoplasmic granular and filamentous aggregates of similar density and continuous with the muscle Z discs (Fig. 1). In some regions, abnormal sarcomeres were seen with no clear demarcation of I and A bands. Disintegrated fibers were prevalent, with sparse filaments in some areas and clumps or aggregates of structural material in others. Individual muscle fibers often were misaligned and/or disorganized. Splitting of myofibrils also was observed, perturbing the Z band registry. Similar disruptions of myofibrils and aggregations of desmin filaments were seen in the myocardium and intestinal smooth muscle (18). These are the classical features of desminopathies (16, 26, 27), and similar abnormalities have been observed in desmin null mutant mice (12–15). In the mice, myofibrils are fragile upon mechanical stress, and muscle weakness develops with age. This mechanosensitive aspect of the phenotype was similar in our patient, who developed muscle weakness in his teens.
Figure 1.
Ultrastructural abnormalities in a skeletal muscle biopsy. Note splitting of myofibrils and aberrations in Z band register. Subsarcolemmal aggregates of granular and filamentous material were prevalent. The aggregates show the same electron density as the Z bands, and are in continuity with them. Similar ultrastructural images were seen in myocardium and in small intestine muscularis propria (18). (Bar represents 1 μm.)
A Mutation Encoding the Deletion of R173 to E179 in the Helix 1B Domain of Desmin and the Lack of Wild-Type Desmin.
Given the similarities between the desmin aggregates in the muscles of our patient and the keratin aggregates that exist in severely affected epidermolysis bullosa simplex cases, the possibility was examined that one or more of the desmin alleles might be altered in this patient. SSCP was used with PCR and exon-specific primers (17) to amplify the desmin coding sequences and splice sites from genomic DNAs of the patient and several normal individuals. Sets of amplified exons then were denatured, placed on ice, and subjected to nondenaturing gel electrophoresis.
As shown in the first lane of Fig. 2B, the coding and noncoding strands from the second half of exon 1 of the wild-type desmin gene could be readily separated under these gel conditions. Additional wild-type samples also displayed this pattern, suggesting that if a polymorphism existed within this segment of the desmin gene, it did not resolve from the wild-type sequence under the conditions used. In contrast, the SSCP pattern of the corresponding portion of desmin exon 1 from the patient DNA was very different from wild type (second lane in Fig. 2B). Both the coding and the noncoding strands migrated aberrantly in repeated (>3) independent PCR/SSCP analyses of this patient’s DNA. That the wild-type coding and noncoding strands were missing from the patient was surprising, because a heterozygous change in the genome still would have left one wild-type allele to generate these strands.
Figure 2.
(A) Pedigree of the desmin-related myopathy family. Family members from whom DNA samples were obtained are numbered. Squares: males. Circles: females. Solid symbol: patient. Open symbols: wild-type individuals. Semiopen symbols: carriers of the deletion. Crossed symbol: deceased individuals. (B) SSCP analysis showing the different single-strand band pattern of exon 1 (4FR) amplimers from the proband (II.1) and a wild-type control individual (WT). (C) Exon 1 amplimers from the available family members and a control individual (WT) run in a denaturing acrylamide gel to detect wild-type (437 nt) and mutant (416 nt) alleles.
To assess the nature of the abnormality detected by SSCP, the PCR products from the patient’s exon 1 reaction were cloned and sequenced. Within exon 1, there was a 21-bp deletion spanning nucleotide positions 679–699. This deletion predicted a desmin protein lacking amino acid residues R173–E179 within the coiled-coil segment of helix 1B (Fig. 3). This deletion was observed in 100% of the clones of exon 1 from the patient. Taken together with the absence of normal allelic bands in the SSCP pattern, this finding suggested that the patient was homozygous for the alteration.
Figure 3.
Location of the putative deletion in relation to desmin’s secondary structure. IF proteins share a common secondary structure consisting of four α-helical, coiled-coil rod segments 1A, 1B, 2A, and 2B, separated by short nonhelical linker sequences (thin lines) and flanked by nonhelical head and tail segments (29). The human desmin amino acid sequence corresponding to the end of helix 1A (amino acids 133–143), linker L1 (amino acids 144–151), and the beginning of helix 1B (amino acids 152–181) are shown. The A135 (see text) and the seven deleted residues (173–179) are highlighted (28). Arrow indicates approximate localization of the deletion.
We next turned to the DNAs from available family members. This time, the PCR-amplified segments of exon 1 were examined by using electrophoretic conditions that resolved the wild-type from the 21-bp deletion bands in their single-stranded state. As shown in Fig. 2 A and C, the patient’s mother and two of his siblings were heterozygous for the mutation, whereas a sister was wild type. The offspring from a heterozygous sibling was also wild type. Because DNA from the father was not available, we cannot discard the possibility that the proband was hemizygous for this allele. Finally, exon 1 from 100 wild-type desmin alleles behaved as the representative wild-type samples shown here. Thus, this genetic alteration appeared to be specific for this family and could not be ascribed to a common sequence polymorphism present in the normal population. Because none of the heterozygous carriers have yet to display major signs of generalized myopathy, the clinical features of the patient correlated with the loss of the wild-type desmin allele and the presence of the mutant allele.
In all control samples analyzed and in the patient, a 3-bp insertion at nucleotide 568 was detected that differs from the reported sequence (17, 30). This change predicts an additional alanine at residue 135 in helix 1A (Fig. 3). This alanine is also present in hamster desmin (31), and the length of helix 1A predicted from our sequence is conserved among most, if not all, other IF proteins (29). Taken together, it seems likely that the sequence we report corresponds to the human desmin sequence that exists in most people.
Functional Evidence That The 7-aa Residue Deletion Within Helix 1B Is Deleterious to Desmin Filament Network Formation In Vivo.
Previous studies have shown that hamster desmin can form a de novo intermediate filament network in the MCF-7 epithelial cells containing an endogenous IF network composed only of keratin (21). Keratin and desmin IF proteins do not copolymerize, and the desmin network that forms is distinct from the keratin network. In transfected cells, amino or carboxy terminal truncated forms of desmin are unable to form a bona fide IF network on their own, and they disrupt a type III IF network in cell types that express vimentin (20, 21, 32, 33). Moreover, transgenic mice expressing a dominant negative desmin mutant display ultrastructural perturbations in muscles (34).
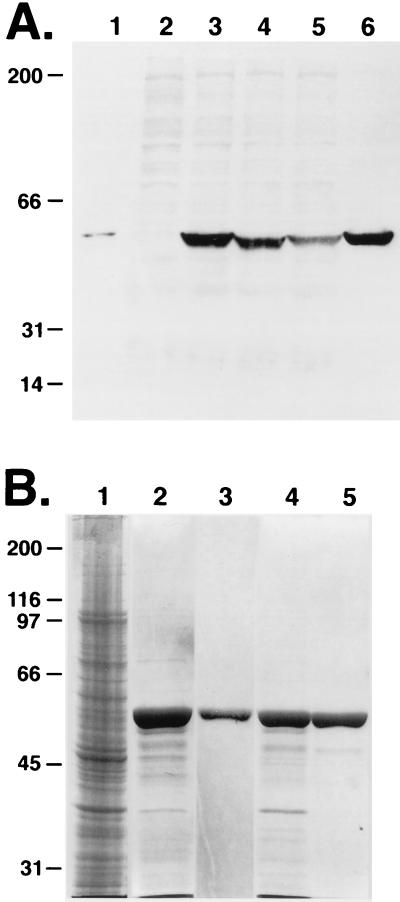
To assess the ability of the Δ173–179 desmin mutation to assemble into a de novo filament network, wild-type and mutant desmin expression vectors were transiently transfected into MCF-7 cells. As determined by antidesmin immunoblot analysis, both vectors generated a single protein of the expected 52-kDa size, with mutant desmin migrating only slightly faster than wild type (Fig. 4A). These bands also were detected in COS simple epithelial cells transfected with the desmin expression vectors. Studies using a similar vimentin expression vector and an antivimentin antibody confirmed that although MCF-7 cells do not express endogenous vimentin, transiently expressed vimentin is stable and exists as a single band of 57 kDa (not shown; see refs. 21 and 22). These data verified that the mammalian expression vectors engineered for these studies generated stable proteins at comparable levels.
Figure 4.
(A) Antidesmin immunoblot analysis of protein extracts. Proteins were from mouse heart muscle (lane 1), MCF-7 cells transfected with: pCMV (lane 2), pCMVDes (lanes 3 and 5), pCMVΔ7Des (lane 4), pJVim (lane 5), or COS cells transfected with pCMVDes (lane 6). (B) SDS/PAGE analyses of bacterially expressed desmin proteins. Escherichia coli strain BL21(DE3)pLysS transformed with pETDes or pETΔ7Des were harvested and inclusion body fractions (IBFs) were isolated and used for purification of recombinant desmin proteins by Mono-Q anion exchange chromatography. Protein samples were resolved by SDS/PAGE and visualized by Coomassie blue stain. Samples are from: lane 1, total bacterial extract (from pETDes transformed cells); lanes 2 and 4, IBF insoluble protein extracts from pETDes or pETΔ7Des-transformed cells; lanes 3 and 5, purified Des or Δ7Des after Mono-Q anion exchange chromatography. Molecular mass standards are at left in kDa.
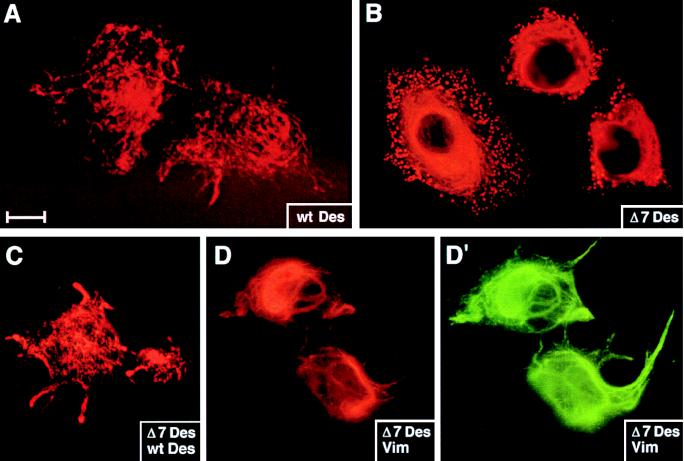
Immunofluorescence microscopy was used to assess the ability of these proteins to produce IF networks in MCF-7 cells. Antidesmin antibody staining was detected in ≈50% of all transfected cells, and not in any untransfected or mock-transfected populations. Wild-type desmin produced an extensive filament network (Fig. 5A), in a fashion similar to that observed previously with a desmin expression vector containing the rous sarcoma virus promoter (21, 22). Removal of seven amino acid residues from helix 1B had a deleterious effect on the ability of desmin to form a normal IF network in vivo. Thus, more than 50% of the cells transfected with the Δ173–179 desmin mutant (Δ7Des) displayed a perinuclear array of antidesmin labeling fibers with clumps or aggregates of desmin at the cell periphery (examples shown). Other transfected cells exhibited only aggregates of antidesmin positive material, with little or no discernable fiber-like network. Very few (<10%) of the transfected cells displayed a normal-looking network of desmin filaments.
Figure 5.
De novo desmin IF network formation in MCF-7 cells. Cells were transfected with pCVM (not shown), pCMVDes (A and C), pCMVΔ7Des (B–D′), or pJVim (D and D′). After transient expression of the transgenes, cells were fixed and stained with antibodies against desmin (A–C and D′) or vimentin (D). Shown are representative examples of transfected cells. Under these conditions, no staining was observed in pCVM-transfected or in untransfected cells. (Bar represents 15 μm in A and B, 20 μm in C–D′.)
Because family members harboring the heterozygous mutation did not display clinical signs of severe myopathy, it was important to examine the behavior of the Δ173–179 desmin mutant in a situation where wild-type desmin also was expressed. As shown in Fig. 5C, a number of MCF-7 cells cotransfected with both wild-type and mutant desmin expression vectors displayed a seemingly normal desmin filament network. Such networks were typical of ≈50% of cotransfected cells. Nearly all of the cotransfected cells displayed some antidesmin-labeled filaments. Only ≈20% of cotransfected cells produced signs of punctate antidesmin staining, the feature so prominent in cells transfected with the mutant construct alone.
Vimentin is not normally expressed in adult muscle nor is it induced in mice lacking desmin (12–14). Consistent with these observations is the absence of antivimentin staining in adult muscle tissue of this patient (18). This said, it is well established that during embryogenesis, vimentin is coexpressed with desmin in developing muscles (1), and therefore it was important to assess the behavior of the Δ173–179 desmin mutant in the presence of vimentin. As shown in Fig. 5D, mutant desmin coassembled into a filamentous network with vimentin. The behavior of the Δ173–179 desmin mutant in the presence of vimentin was similar to that observed with a mixture of wild-type and mutant desmin. Taken together, these findings suggest that during embryonic development the effect of the mutant desmin in our patient was masked by the presence of vimentin.
The Δ173–179 Mutation Severely Compromises the Ability of Desmin to Form 10-nm Filaments in Vitro.
To further examine the consequences of the Δ173–179 desmin mutation on IF assembly, we used a bacterial expression system to overproduce mutant and wild-type desmin (24). After purification, recombinant proteins first were examined by SDS/PAGE. As judged by densitometry analysis of the Coomassie blue-stained gels, the proteins were judged to be >98% pure (Fig. 4B). These proteins then were reconstituted in an in vitro filament assembly assay (25).
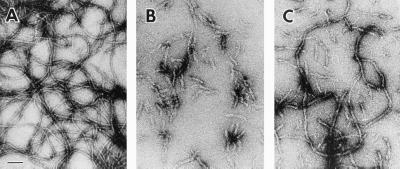
Purified wild-type desmin assembled very efficiently into long IFs (Fig. 6A). These filaments were uniform in diameter (9.8 nm ± 0.1 nm) and in length (5–10 μm), and at the concentrations used, they readily formed viscous gels, reflective of their filament network forming ability (see ref. 29 for review).
Figure 6.
In vitro assembly of wild-type and mutant desmin. Proteins were purified and subjected to in vitro filament assembly (see Materials and Methods). (A) Desmin. (B) Δ7 Desmin. (C) 1:1 mixture of desmin +Δ7 desmin. (Bar represents 100 nm.)
In contrast, the Δ173–179 desmin mutant formed short, wide, and irregular rod-like filaments (Fig. 6B). The average width of these structures was 15 nm ± 0.5 nm, considerably larger than the standard ≈10-nm diameter of IFs. The greater dispersion around the mean was reflective of the heterogeneity in the structures generated by this mutant. Although not quantitated formally, filaments ranged from 80 to 450 nm; most were shorter than 250 nm and exhibited a tendency to form loose aggregates. This said, the polymerization efficiency was high (see Materials and Methods), confirming the ability of the mutant desmin to associate into these aberrant fibrous structures. The assembly behavior of mutant desmin did not improve even at concentrations as high as 500 μg/ml.
Mixtures of wild-type and mutant desmin displayed an intermediate behavior, depending on the stoichiometry of the two proteins. A 1:1 molar ratio resulted in the formation of filaments, which were longer than those seen in assembly reactions with mutant alone (Fig. 6C). These filaments were more similar, but still larger in diameter to wild-type desmin IFs. In addition, many short filaments were still observed, indicating that assembly was improved but not fully restored in a situation more analogous to the heterozygous state. A 1:5 molar ratio in favor of wild-type desmin was necessary to form reasonably long (>1 μm) filaments of near normal diameter; however, even at this ratio, the filaments were significantly more irregular than the wild-type control (data not shown).
DISCUSSION
The discovery of a homozygous or hemizygous mutation in the desmin gene of our patient marks a case in which a desmin mutation has been associated with a generalized myopathy in humans. The mutation resides in the beginning of the helix 1B within the central α-helical rod segment of desmin. The predicted polypeptide of our patient’s desmin lacks a heptad of hydrophobic residues within this domain. These heptad repeats of hydrophobic residues, where most a and d residues of each a b c d e f g peptide are hydrophobic, form the underlying basis for coiled-coil formation of IF dimer subunits (29). Although the deletion of precisely seven amino acid residues does not change the periodicity of the coiled-coil interaction, it might affect the ability of dimer subunits to form functional higher order structures. In this regard, it may be relevant that the lengths of the four α-helical segments are highly evolutionarily conserved among IF proteins, and of the IF superfamily, only the nuclear lamins have a longer helix 1B (for review, see ref. 29).
Our functional studies provide compelling evidence that the deletion of a heptad within helix 1B is deleterious to desmin filament assembly both in vivo and in vitro. Interestingly, whereas the mutation severely affected the ability of desmin to assemble into bona fide 10-nm filaments, short rodlets of aberrantly large diameter assembled in vitro, and these rodlets displayed a strong tendency to aggregate. This finding may explain at least in part why desmin network formation was appreciable, although clearly aberrant, in cultured cells transfected with the mutant desmin transgene. This said, the effects of the desmin mutation seemed to be more severe in vitro than in vivo. Whether this difference is because of more optimal buffer or ionic conditions in vivo or to as yet unidentified cofactor proteins that might enhance IF assembly remains to be determined. In any case, the deleterious consequences of this mutation to filament assembly seem likely to be responsible for the muscle derangements observed in this patient.
It is interesting that filament assembly was dramatically improved in the presence of wild-type desmin or vimentin. It seems unlikely that improvement stems from interactions at the level of the dimer, because a heterodimer would be expected to perturb the alignment within the coiled-coil. Although such interactions may occur, we anticipate that the beneficial action of the wild-type IF protein comes from a higher ordered level of association. The improvement on filament network formation was most striking in vivo, where a number of cells formed type III IF networks that were nearly indistinguishable from wild type. This finding may explain why no major clinical signs of myopathy occurred in the heterozygous carriers within the family. Whether there might be more subtle muscle defects in the family members who harbor a single copy of the mutant allele is an issue that has thus far been precluded by our inability to obtain a muscle biopsy.
Despite the promising correlation between the loss of a desmin network and muscle degeneration in knockout mice, the human desmin gene at chromosome band 2q35 recently was excluded as a locus involved in desmin-related myopathy in three different families (17). However, this exclusion does not necessarily rule out the possibility that certain types of desminopathies may harbor desmin gene defects, because myopathies associated with desmin abnormalities are highly heterogeneous. They include cardiomyopathies, congenital myopathies with either childhood or adult onset of disease, cardiomyopathy associated with myopathy, and a multisystemic disorder involving skeletal muscle, heart, peripheral nerves, and intestinal muscularis propria (16, 35–38). In addition, morphological abnormalities in desmin-related myopathies are diverse, as is their mode of inheritance. Thus, when taken together with the functional desmin gene knockout data in mice, it is not surprising that despite the negative results of Vicart et al. (17), we have found a mutation in the desmin gene of a patient with generalized muscle degeneration.
Our functional studies suggest that the mutation is causative, and that the irregularities seen in the alignment of myofibrils and Z bands and the disorganization and splitting of myofibrils in the muscles of this patient can be attributed to primary perturbations in the desmin filament architecture at the Z bands. The decline in muscle integrity with age in this patient correlates well with the notion that exercise and repeated muscle contractions test the mechanical and tensile strength of the myofibrillar Z bands, where desmin IFs are thought to function. In the absence of wild-type desmin, the desmin mutation harbored by this patient would be expected to severely impair, if not obliterate, desmin function.
There are still some aspects of the clinical features of the patient and of the pathology that we cannot attribute to a mutation in desmin. In particular was the congenital sensory deafness of the patient and defects in postmortem nerve tissue, including a slight thinning of myelin sheaths in cranial and peripheral nerves, and axonal spheroids in the spinal cord and dorsal roots (18). Because the heterozygous brother has developed deafness in the absence of any apparent muscle abnormalities, the two features do not appear to be related. The nerve abnormalities are likely effects of vitamin malabsorption caused by defective bowel smooth muscle function.
Irrespective of these issues, our identification of a desmin gene deletion that functionally compromises desmin IF assembly in this patient underscores the significance of desmin gene alterations in the production of desmin-related muscle pathology in humans. From these findings, we surmise that additional generalized myopathies exhibiting ultrastructural alterations in desmin will arise from mutations in the human desmin gene. Given the discovery of both dominant negative and recessive null mutations in keratin IF disorders (5), such myopathies could be autosomal dominant or recessive in nature. Based on the mutations identified in keratin IF disorders (5), the majority of autosomal dominant desmin mutations are predicted to reside at the ends of the α-helical coiled-coil rod segment rather than at the more internal location observed in our patient, who was hemizygous or homozygous for the desmin mutation.
Acknowledgments
We are grateful to J. Pérez-Piteira (Department of Pathology, Hospital Universitari Germans Trias i Pujol) and to G. Marfany, A. Martínez, M. Bayés, and B. Cormand (Department of Genetics, Facultat de Biologia, Univ. de Barcelona) for stimulating discussion and expert technical advice. We thank Dr. P. Vicart and Dr. D. Paulin (Institut Pasteur, Paris) for desmin specific primers, Dr. Hans Bloemendal (Univ. of Nijmegen, The Netherlands) for his gift of the hamster desmin cDNA clone, and Dr. Christoph Bauer and Chuck Wellek for their assistance in preparing figures for this paper. This research was supported by Comision Interministerial de Ciencia y Tecnologia Grants SAF 97/0239 and SAF 97/0227 of the Spanish Ministries of Health and Education, Fundacio Marató TV3 Pi i Sunyer 3215/97 and 1033/97, and by grants from the National Institutes of Health (R01-AR27883 to E.F.; R01-AR42047 to P.C.). X.R. is recipient of a Comisio Interdepartamental de Recerca i Innovacio Tecnologica-FI predoctoral fellowship. A.M.M.-M. and E.V. are recipients of a fellowship from Fundació Echevarne. E.F. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- IF
intermediate filament
- CMV
cytomegalovirus
- SSCP
single-stranded conformation polymorphism
Note Added in Proof
While this manuscript was in press, Goldfarb et al. (39) reported a heterozygous A337P (adult onset) and a compound heterozygous A360P/N393I substitution (childhood onset) in the desmin allele(s) of two generalized myopathy cases. No functional studies were conducted to assess whether these substitutions are causative.
References
- 1.Li Z, Marchand P, Humbert J, Babinet C, Paulin D. Development (Cambridge, UK) 1993;117:947–959. doi: 10.1242/dev.117.3.947. [DOI] [PubMed] [Google Scholar]
- 2.Gard D L, Lazarides E. Cell. 1980;19:263–275. doi: 10.1016/0092-8674(80)90408-0. [DOI] [PubMed] [Google Scholar]
- 3.Lazarides E, Granger B L, Gard D L, O’Connor C M, Breckler J, Price M, Danto S I. Cold Spring Harbor Symp Quant Biol. 1982;46:351–378. doi: 10.1101/sqb.1982.046.01.036. [DOI] [PubMed] [Google Scholar]
- 4.Lazarides E, Hubbard B D. Proc Natl Acad Sci USA. 1976;73:4344–4348. doi: 10.1073/pnas.73.12.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs E, Cleveland D. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- 6.Vassar R, Coulombe P A, Degenstein L, Albers K, Fuchs E. Cell. 1991;64:365–380. doi: 10.1016/0092-8674(91)90645-f. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd C, Yu Q-C, Cheng J, Turksen K, Degenstein L, Hutton E, Fuchs E. J Cell Biol. 1995;129:1329–1344. doi: 10.1083/jcb.129.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulombe P A, Hutton M E, Letai A, Hebert A, Paller A S, Fuchs E. Cell. 1991;66:1301–1311. doi: 10.1016/0092-8674(91)90051-y. [DOI] [PubMed] [Google Scholar]
- 9.Bonifas J M, Rothman A L, Epstein E H. Science. 1991;254:1202–1205. doi: 10.1126/science.1720261. [DOI] [PubMed] [Google Scholar]
- 10.Lane E B, Rugg E L, Navsaria H, Leigh I M, Heagerty A H M, Ishida-Yamamoto A, Eady R A J. Nature (London) 1992;356:244–246. doi: 10.1038/356244a0. [DOI] [PubMed] [Google Scholar]
- 11.Letai A, Coulombe P A, McCormick M B, Yu Q-C, Hutton E, Fuchs E. Proc Natl Acad Sci USA. 1993;90:3197–3201. doi: 10.1073/pnas.90.8.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Colucci-Guyon E, Pincon-Raymond M, Mericskay M, Pournin S, Paulin D, Babinet C. Dev Biol. 1996;175:362–363. doi: 10.1006/dbio.1996.0122. [DOI] [PubMed] [Google Scholar]
- 13.Milner D J, Weitzer G, Tran D, Bradley A, Capetanaki Y. J Cell Biol. 1996;134:1255–1270. doi: 10.1083/jcb.134.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Mericskay M, Agbulut O, Butler-Browne G, Carlsson L, Thornell L E, Babinet C, Paulin D. J Cell Biol. 1997;139:129–144. doi: 10.1083/jcb.139.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sjuve R, Arner A, Li Z, Mies B, Paulin D, Schmittner M, Small J V. J Muscle Res Cell Motil. 1998;19:415–429. doi: 10.1023/a:1005353805699. [DOI] [PubMed] [Google Scholar]
- 16.Goebel H H, D’Agostino A N, Wilson J, Cole G, Foroud T, Koller D, Farlow M, Azzarelli B, Muller J. Muscle Nerve. 1997;20:1127–1136. doi: 10.1002/(sici)1097-4598(199709)20:9<1127::aid-mus6>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 17.Vicart P, Dupret J M, Hazan J, Li Z, Gyapay G, Krishnamoorthy R, Weissenbach J, Fardeau M, Paulin D. Hum Genet. 1996;98:422–429. doi: 10.1007/s004390050233. [DOI] [PubMed] [Google Scholar]
- 18.Ariza A, Coll J, Fernandez-Figueras T, Lopez M D, Mate J L, Garcia O, Fernandez-Vasalo A, Navas-Palacios J J. Hum Pathol. 1995;26:1032–1037. doi: 10.1016/0046-8177(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 19.Bayes M, Valverde D, Balcells D, Grinberg D, Vilageliu L, Benitez J, Ayuso C, Beneyto M, Baiget M, Gonzalez-Duarte R. Hum Genet. 1995;96:89–94. doi: 10.1007/BF00214192. [DOI] [PubMed] [Google Scholar]
- 20.Raats J M H, Pieper F R, Vree Egberts W T M, Verrijp K N, Ramaekers F C S, Bloemendal H. J Cell Biol. 1990;111:1971–1985. doi: 10.1083/jcb.111.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raats J M H, Henderik J B J, Verdijk M, van Oort F L G, Gerards W L H, Ramaekers F C S, Bloemendal H. Eur J Cell Biol. 1991;56:84–103. [PubMed] [Google Scholar]
- 22.McCormick M B, Kouklis P, Syder A, Fuchs E. J Cell Biol. 1993;122:395–407. doi: 10.1083/jcb.122.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studier F W, Moffatt B A. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 24.Coulombe P, Fuchs E. J Cell Biol. 1990;111:153–169. doi: 10.1083/jcb.111.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrmann H, Hoffmann I, Franke W W. J Mol Biol. 1992;223:637–650. doi: 10.1016/0022-2836(92)90980-x. [DOI] [PubMed] [Google Scholar]
- 26.Pellissier J F, Pouget J, Charpin C, Figarella D. J Neurol Sci. 1989;89:49–61. doi: 10.1016/0022-510x(89)90006-3. [DOI] [PubMed] [Google Scholar]
- 27.Goebel H H. Curr Opin Neurol. 1997;10:426–429. doi: 10.1097/00019052-199710000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Geisler N, Weber K. Eur Mol Biol Organ J. 1982;1:1649–1656. [Google Scholar]
- 29.Fuchs E, Weber K. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 30.Li Z L, Lilienbaum A, Butler-Browne G, Paulin D. Gene. 1989;78:243–254. doi: 10.1016/0378-1119(89)90227-8. [DOI] [PubMed] [Google Scholar]
- 31.Quax W, Egberts W V, Hendriks W, Quax-Jeuken Y, Bloemendal H. Cell. 1983;35:215–223. doi: 10.1016/0092-8674(83)90224-6. [DOI] [PubMed] [Google Scholar]
- 32.Raats J M, Gerards W L, Schreuder M I, Grund C, Henderik J B, Hendriks I L, Ramaekers F C, Bloemendal H. Eur J Cell Biol. 1992;58:108–127. [PubMed] [Google Scholar]
- 33.Yu K R, Hijikata T, Lin Z X, Sweeney H L, Englander S W, Holtzer H. Proc Natl Acad Sci USA. 1994;91:2497–2501. doi: 10.1073/pnas.91.7.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raats J M H, Schaart G, Henderik J B J, van der Kemp A, Dunia I, Benedetti E L, Pieper F R, Ramaekers F C S, Bloemendal H. Eur J Cell Biol. 1996;71:221–236. [PubMed] [Google Scholar]
- 35.Cary R B, Klymkowsky M W. Differentiation. 1994;56:31–38. doi: 10.1046/j.1432-0436.1994.56120031.x. [DOI] [PubMed] [Google Scholar]
- 36.Fardeau M, Godet-Guillain J, Tome F M, Collin H, Gaudeau S, Boffety C, Vernant P. Rev Neurol. 1978;134:411–425. [PubMed] [Google Scholar]
- 37.Pellissier J F, Pouget J, Charpin C, Figarella D. J Neurol Sci. 1989;89:49–61. doi: 10.1016/0022-510x(89)90006-3. [DOI] [PubMed] [Google Scholar]
- 38.Baeta A M, Figarella-Branger D, Bille-Turc F, Lepidi H, Pellissier J F. Acta Neuropathol. 1996;92:499–510. doi: 10.1007/s004010050552. [DOI] [PubMed] [Google Scholar]
- 39.Goldfarb L G, Park K Y, Cervenakova L, Gorokhova S, Lee H S, Vasconcelos O, Nagle J W, Semino-Mora C, Sivakumar K, Dalakas M C. Nat Genet. 1998;19:402–403. doi: 10.1038/1300. [DOI] [PubMed] [Google Scholar]