Abstract
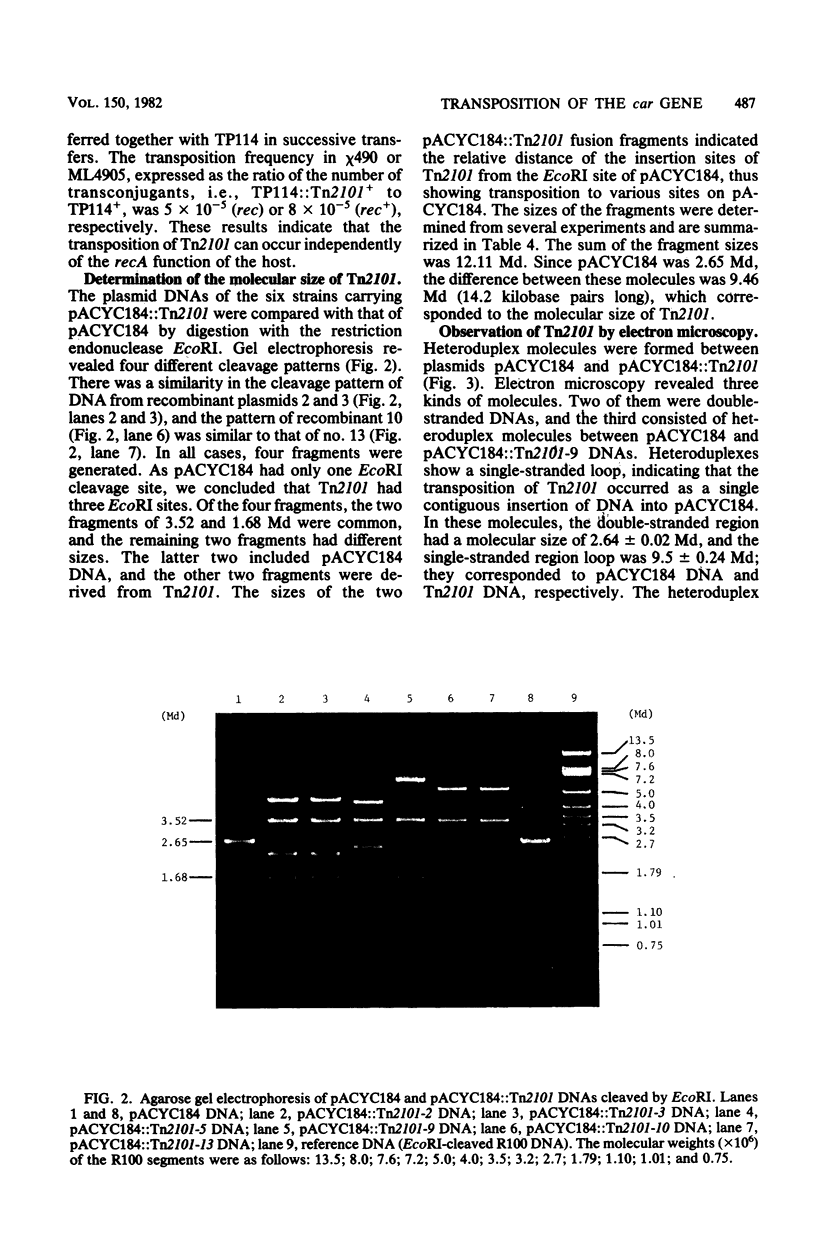
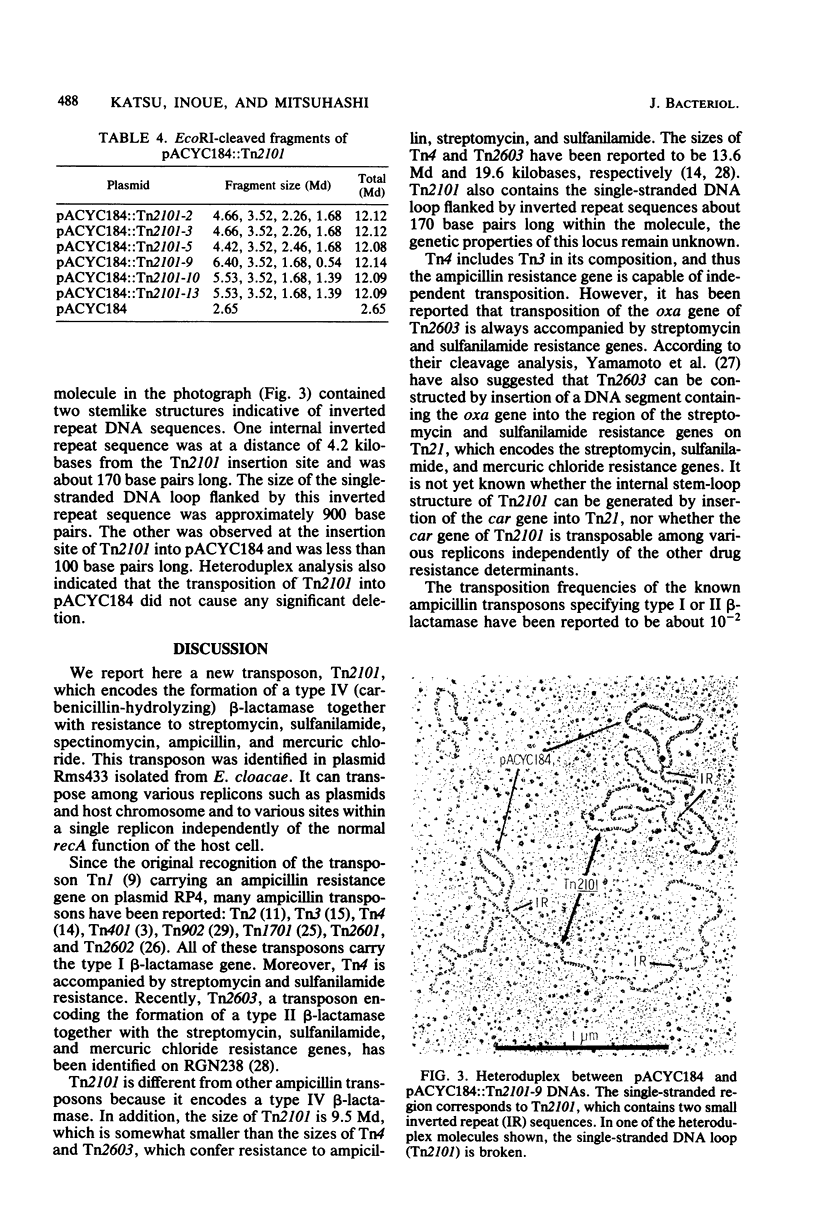
We isolated a new transposon Tn2101, from plasmid Rms433 in Enterobacter cloacae. Tn2101 encoded the formation of type IV (carbenicillin-hydrolyzing) beta-lactamase and multiple resistance to streptomycin, sulfanilamide, spectinomycin, and mercury in addition to ampicillin. Tn2101 was transposable between conjugative (or nonconjugative) plasmids and the host chromosome. Transposition occurred independently of the general recombination ability of the host cell. Tn2101 had a molecular size of 9.5 x 10(6) and contained short inverted repeat terminal sequences.
Full text
PDF

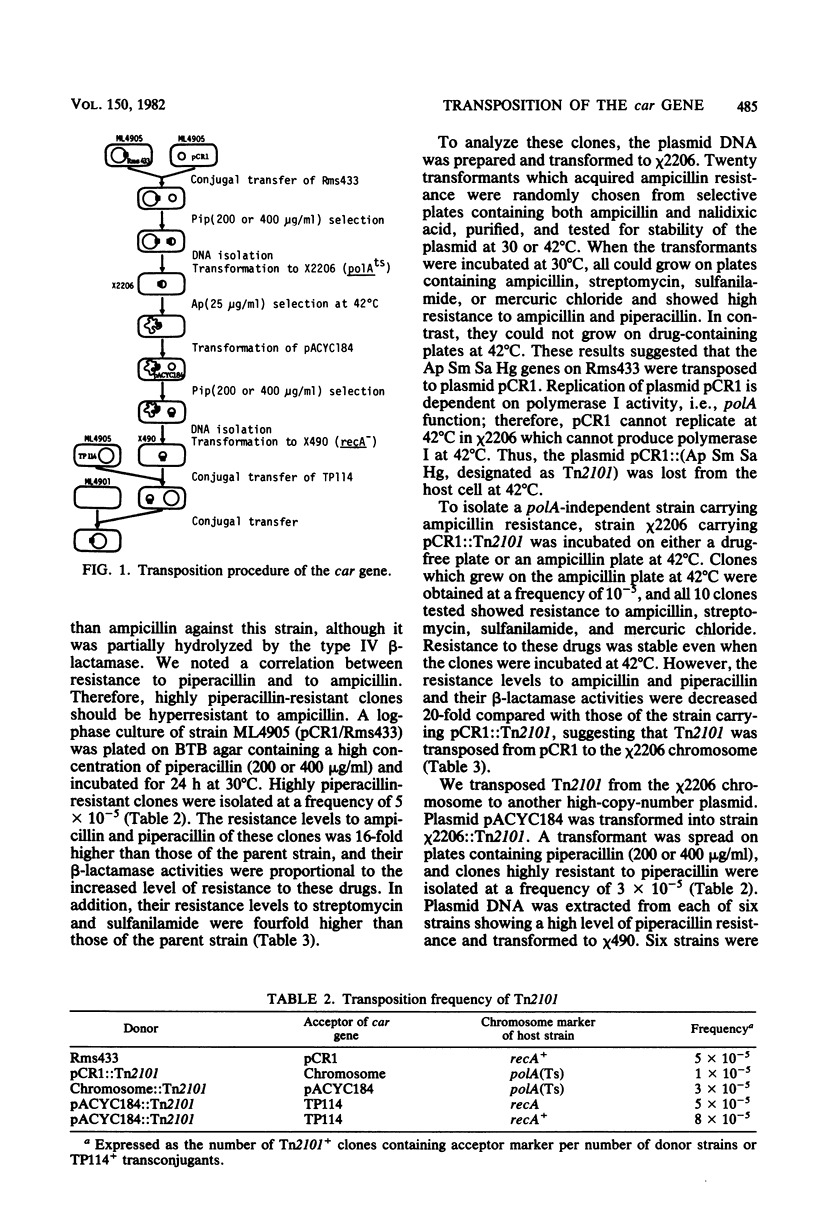
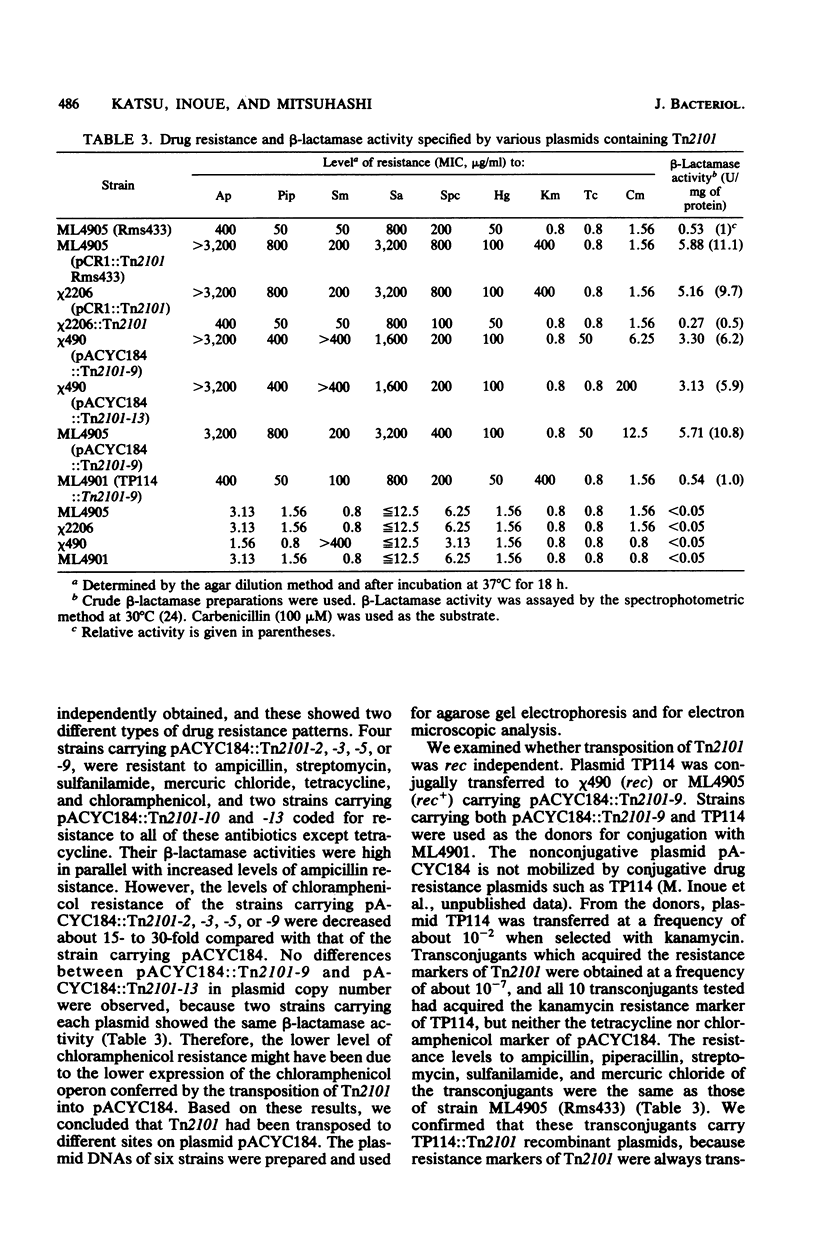

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K. A., Hershfield V., Helinski D. R. Gene cloning and containment properties of plasmid Col E1 and its derivatives. Science. 1977 Apr 8;196(4286):172–174. doi: 10.1126/science.322277. [DOI] [PubMed] [Google Scholar]
- Beck E., Sommer R., Auerswald E. A., Kurz C., Zink B., Osterburg G., Schaller H., Sugimoto K., Sugisaki H., Okamoto T. Nucleotide sequence of bacteriophage fd DNA. Nucleic Acids Res. 1978 Dec;5(12):4495–4503. doi: 10.1093/nar/5.12.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedik M., Fennewald M., Shapiro J. Transposition of a beta-lactamase locus from RP1 into Pseudomonas putida degradative plasmids. J Bacteriol. 1977 Feb;129(2):809–814. doi: 10.1128/jb.129.2.809-814.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Grindley J. N., Anderson E. S. R factor compatibility groups. Mol Gen Genet. 1972;119(4):287–297. doi: 10.1007/BF00272087. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Datta N., Kontomichalou P., Smith J. T. Molecular specificities of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol. 1974 Jan;117(1):56–62. doi: 10.1128/jb.117.1.56-62.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Matthew M. Acquisition by Escherichia coli of plasmid-borne beta-lactamases normally confined to Pseudomonas spp. Plasmid. 1979 Apr;2(2):269–278. doi: 10.1016/0147-619x(79)90045-3. [DOI] [PubMed] [Google Scholar]
- Heffron F., Sublett R., Hedges R. W., Jacob A., Falkow S. Origin of the TEM-beta-lactamase gene found on plasmids. J Bacteriol. 1975 Apr;122(1):250–256. doi: 10.1128/jb.122.1.250-256.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike Y., Hashimoto H., Motohashi K., Fujisawa N., Mitsuhashi S. Isolation and characterization of a composite plasmid Rms201 mutant temperature sensitive for replication. J Bacteriol. 1980 Feb;141(2):577–583. doi: 10.1128/jb.141.2.577-583.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsu K., Inoue M., Mitsuhashi S. Plasmid-mediated carbencillin hydrolyzing beta-lactamases of Proteus mirabilis. J Antibiot (Tokyo) 1981 Nov;34(11):1504–1505. doi: 10.7164/antibiotics.34.1504. [DOI] [PubMed] [Google Scholar]
- Kopecko D. J., Brevet J., Cohen S. N. Involvement of multiple translocating DNA segments and recombinational hotspots in the structural evolution of bacterial plasmids. J Mol Biol. 1976 Dec;108(2):333–360. doi: 10.1016/s0022-2836(76)80124-6. [DOI] [PubMed] [Google Scholar]
- Kopecko D. J., Cohen S. N. Site specific recA--independent recombination between bacterial plasmids: involvement of palindromes at the recombinational loci. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1373–1377. doi: 10.1073/pnas.72.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupersztoch-Portnoy Y. M., Lovett M. A., Helinski D. R. Strand and site specificity of the relaxation event for the relaxation complex of the antibiotic resistance plasmid R6K. Biochemistry. 1974 Dec 31;13(27):5484–5490. doi: 10.1021/bi00724a005. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Matthew M., Hedges R. W. Analytical isoelectric focusing of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol. 1976 Feb;125(2):713–718. doi: 10.1128/jb.125.2.713-718.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanak N., Cramer J. H., Rownd R. H. EcoRI restriction endonuclease map of the composite R plasmid NR1. J Bacteriol. 1976 Jul;127(1):619–636. doi: 10.1128/jb.127.1.619-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Nagai Y., Hashimoto H., Mitsuhashi S. Distribution of R factors among Shigella strains isolated in Japan. Jpn J Microbiol. 1969 Jun;13(2):187–191. doi: 10.1111/j.1348-0421.1969.tb00452.x. [DOI] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid. 1977 Nov;1(1):1–7. doi: 10.1016/0147-619x(77)90003-8. [DOI] [PubMed] [Google Scholar]
- Waley S. G. A spectrophotometric assay of beta-lactamase action on penicillins. Biochem J. 1974 Jun;139(3):789–790. doi: 10.1042/bj1390789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Calame K. L., Grindley J. N., Nakada D. Location of an ampicillin resistance transposon, Tn1701, in a group of small, nontransferring plasmids. J Bacteriol. 1979 Feb;137(2):990–999. doi: 10.1128/jb.137.2.990-999.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Katoh R., Shimazu A., Yamagishi S. Gene expression of ampicillin resistance transposons, Tn2601 and Tn2602. Microbiol Immunol. 1980;24(6):479–494. doi: 10.1111/j.1348-0421.1980.tb02852.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Baba R., Yamagishi S. Physical and functional mapping of Tn2603, a transposon encoding ampicillin, streptomycin, sulfonamide, and mercury resistance. Mol Gen Genet. 1981;181(4):464–469. doi: 10.1007/BF00428737. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Nohara C., Fukunaga Y., Yamagishi S. Transposition of the oxacillin-hydrolyzing penicillinase gene. J Bacteriol. 1981 Feb;145(2):808–813. doi: 10.1128/jb.145.2.808-813.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]