Abstract
The envelope (Env) proteins of primate lentiviruses interact sequentially with CD4 and a coreceptor to infect cells. Changes in coreceptor use strongly influence viral tropism and pathogenesis. We followed the evolution of coreceptor use in pig-tailed macaques that developed severe CD4 T-cell loss during the derivation of a pathogenic simian HIV (SHIV) that contained the tat, rev, vpu, and env genes of the HXBc2 strain of HIV-1 in a genetic background of SIVmac239. The Env from the parental virus as well as one derived from the first macaque to develop AIDS exclusively used CXCR4 as a coreceptor, indicating that CXCR4 can function as a coreceptor in macaques even though it is rarely used by simian immunodeficiency viruses. One Env (Pnb5), obtained from a macrophage-tropic virus isolated from the cerebral spinal fluid, did not use CCR5 or CXCR4. Instead, it used CCR2b and to a lesser extent CCR3, STRL33, and APJ to infect cells. Chimeras between Pnb5 and the parental X4 Env indicated that the V3 loop is the major determinant of CXCR4 use, with other regions of Env influencing the efficiency with which this coreceptor was used. In contrast, the Pnb5 V1/2 and V3 regions in combination were both necessary and sufficient to confer full use of CCR2b, CCR3, STRL33, and APJ to the parental X4 Env protein. These results are consistent with a single, conserved binding site in Env that interacts with multiple coreceptors in conjunction with the V1/2 and V3 loops, and suggest that the V1/2 region plays a more important role in governing the use of CCR2b, CCR3, STRL33, and APJ than for CXCR4.
Infection of cells by HIV type 1 (HIV-1) involves interactions between the viral envelope (Env) protein, CD4, and a seven transmembrane domain coreceptor (reviewed in refs. 1–5). R5 strains of HIV-1, which are largely responsible for virus transmission, use the chemokine receptor CCR5 to infect cells. Over time, mutations in the viral Env protein can accrue that alter coreceptor dependence and therefore viral tropism. Thus, in approximately 50% of infected individuals, X4 strains of HIV-1 that use CXCR4 can be recovered, though this typically occurs years after infection (6–8). Emergence of X4 virus strains is correlated with accelerated disease progression, indicating that alterations in coreceptor use can strongly influence viral pathogenesis.
Although the selective pressures that drive changes in coreceptor use are poorly understood, the Env determinants responsible for CCR5 and CXCR4 interactions are reasonably well characterized. The most important region in Env that governs CCR5 and CXCR4 use is the V3 loop. Generally, acquisition of basic charges in the V3 loop correlates with CXCR4 use (9–11), perhaps because the regions of CXCR4 most important for coreceptor function are far more acidic than the corresponding regions in CCR5 (12, 13). However, other regions of Env also can influence CCR5 and CXCR4 use, particularly the V1/V2 region. Depending on the overall context in which changes are made, alterations in V1 and V2 sequence alone sometimes can alter viral tropism and coreceptor use (14–18), as can changes in the V3 loop (11, 19–23).
All HIV-1 strains studied to date use CCR5, CXCR4, or both molecules as coreceptors (24). However, nine alternative coreceptors have been identified that support infection by smaller numbers of HIV-1 strains (25–33). Acquisition of the ability to use additional coreceptors may broaden viral tropism and influence disease pathogenesis. CCR3, for example, is expressed in microglia that are the major targets of virus infection in the central nervous system (34). Likewise, the orphan receptor APJ, which can be used by some virus strains to infect cells in vitro, is expressed widely in the brain (30). We have monitored the evolution of coreceptor use over time in pig-tailed macaques infected with a simian HIV (SHIV) containing a X4 HIV-1 Env protein (35–37) and correlated this with changes in viral tropism and Env sequence. Surprisingly, we found that Env proteins derived from multiple SHIV isolates exclusively used CXCR4 as a coreceptor despite the fact that all SIV strains use CCR5 as a coreceptor regardless of tropism (30–32, 38–40). Thus, CXCR4 can function as an efficient coreceptor in macaques even though it is rarely used by simian immunodeficiency virus (SIV). One Env protein (Pnb5), which was isolated from a virus stock (SHIVKU-1), obtained from the cerebral spinal fluid of a pig-tailed macaque that developed severe CD4 T cell loss and AIDS (36), was remarkable for its ability to use CCR2b and to a lesser extent CCR3, STRL33, and APJ to infect cells. The Pnb5 Env differed from the parental X4 Env by 14 amino acid changes, did not use CCR5, used CXCR4 very inefficiently as a coreceptor, and was able to mediate infection of macrophages. Through the construction of chimeric Env proteins, we found that the Pnb5 V3 loop in combination with the V1/V2 region imparted use of CCR2b, CCR3, STRL33, and APJ to the parental HIV-1 HXBc2 Env. Neither the V3 loop or the V1/2 domain alone could impart the ability to use these alternative coreceptors. In contrast, the V3 loop of HXBc2 was both necessary and sufficient to impart use of CXCR4 to the Pnb5 Env, though other domains in Env influenced the efficiency with which this coreceptor was used. Thus, two regions of Env participate in mediating interactions with multiple, diverse HIV-1 coreceptors with the V1/2 region playing a more important role in governing the use of CCR2b, CCR3, STRL33, and APJ than for CXCR4.
MATERIALS AND METHODS
Plasmids.
Human CCR2b, CCR3, CCR5, CCR8, CXCR4, STRL33, GPR15, and CD4 were expressed by using the pcDNA3 vector (Invitrogen), as were rhesus CCR2b, CCR5, and CXCR4. Human GPR1, CX3CR1, and ChemR23 were expressed from pRC/CMV (Invitrogen), pCR3.1 (Invitrogen), and pEFIN3, respectively. To generate plasmids for generating viral pseuodotypes encoding SHIV Envs, the 2,124-bp KpnI–BamHI fragment from the 3′ half of the SHIV-4 and PPc genome was cloned into the corresponding region of psv7d-HXBc2. The 1,208-bp KpnI–BsaBI fragment of gp120 (containing V1-V4 of envelope) from the Pnb5 isolate (37) was cloned into psv7d-SHIV-4 containing the two amino acid changes present in the V5 loop of Pnb5 (N460K and S461G). These V5 substitutions were engineered by using the Quickchange Site-Directed Mutagenesis Kit (Stratagene) and confirmed by DNA sequencing. This work resulted in a full-length Pnb5 Env clone containing amino acid changes from all regions of gp120 in the context of the SHIV-4 Env. Env chimeras between Pnb5 and SHIV-4 envs were generated by using conserved KpnI, PvuII, PpuMI, BsaBI, DraIII, StuI, and Bsu36I sites in both Envs. The identity of all chimeras was confirmed by DNA sequencing. To generate an infectious, replication-competent full-length SHIV virus for macrophage infections containing the Pnb5 Env, the KpnI–BsaBI fragment from Pnb5 was cloned into a plasmid containing the 3′ half of the SHIV-4 genome, which was ligated to the 5′ half of the SHIV-4 genome during production of virus stocks (35).
Passage of SHIV.
The SHIV env genes analyzed in this study were derived from SHIV-4, which contains tat, rev, vpu, and env genes from the HXBc2 strain of HIV-1 in a genetic background of SIVmac239 as previously described (35). A pig-tailed macaque was infected with SHIV-4, and additional animals were infected by sequential in vivo passages of virus (35). The first SHIV-4 passaged env gene analyzed in this study was derived from the spleen tissue from macaque PPc (41), which died from severe CD4+ T cell loss and AIDS at 26 weeks postinoculation (36). The other SHIV-4 derived env was obtained from the uncloned SHIVKU-1 virus, which was derived from subsequent passage of bone marrow cells from macaque PPc and PQc (36, 37). SHIVKU-1 has been shown to cause rapid CD4+ T cell loss and AIDS in pig-tailed macaques.
Luciferase Virus Infection and Protein Expression Studies.
Luciferase reporter viruses were generated by introducing 15 μg of Env plasmid and 5 μg of NL-43 luciferase backbone (pNL-luc-E−R−; refs. 42 and 43) into 293T cells by standard calcium phosphate transfection (using 25-cm2 flasks). The resulting supernatant was centrifuged 2 days posttransfection to remove any contaminating cells and added to target cells. Virus production was found to be similar for each chimeric Env protein as measured by p24 ELISA (NEN Life Science Products). Aliquots of supernatant as well as cell lysate were analyzed by Western blot for the presence of gp160 and gp120 for all Env chimeras. For infection, feline CCCS-CD4 cells were plated in 24-well plates and transfected with 1.5 μg each of additional CD4 and the indicated coreceptor plasmids by using calcium phosphate precipitation. Media was changed on the target cells after 4 hr, which were allowed to express overnight. Target cells were infected the next day with 100 μl of viral supernatant in the presence of 8 μg/ml of DEAE-dextran in a total volume of 500 μl. Strict normalization of virus by p24 content resulted in no qualitative differences in the relative efficiency of coreceptor use between the different Env chimeras. An additional 1 ml of medium was added 1 day after addition of virus to the target cells. Cells were lysed in 150 μl of 0.5% TX-100 3 days postinfection, and 50 μl of the resulting lysate was assayed for luciferase activity.
Infections of Macaque Macrophage Cultures.
Monolayers of macaque macrophage cultures were prepared in 35-mm dishes as previously described (44). Viral stocks of SHIV-4 and Pnb5 were transfected and prepared in C8166 cells, and 1,000 TCID50 (approximately 0.1 ml) was added in 0.5 ml of macrophage differentiation medium (MDM)(44). Cultures were incubated for 2 hr at 37°C, washed three times with MDM, and supplemented with 2 ml of fresh MDM. Macrophage-tropism was assessed by assaying culture supernatants at 0, 4, 6, 8, and 10 days for the presence of p27 core antigen by ELISA. Controls included SHIV-4, which is a lymphocyte-tropic virus, and SHIVKU-1, which is dual-tropic virus strain (37).
RESULTS
Evolution of Coreceptor Use by a Pathogenic SHIV.
A SHIV containing the HXBc2 Env protein on a background of SIVmac239 (SHIV-4) causes persistent infection, but not disease, in pig-tailed macaques (35, 36). Subsequent bone marrow passages in macaques lead to the evolution of a pathogenic SHIV with a limited number of amino acid changes in Env and the ability to replicate in macrophages (36, 37). We have begun to analyze the coreceptors used by pathogenic and nonpathogenic SHIV isolates to determine whether alterations in coreceptor use correlate with changes in pathogenicity and to identify Env determinants responsible for coreceptor interactions. To do this, we tested the coreceptors used by Env proteins obtained from various SHIV isolates by using a luciferase reporter virus system (42, 43). Plasmids expressing the desired Env protein under control of the simian virus 40 promoter were transfected into 293T cells along with a plasmid containing a full-length NL4–3 HIV-1 provirus bearing an inactive Env and luciferase in place of nef. The resulting virus pseudotypes are capable of a single round of replication, resulting in the production of readily quantifiable luciferase activity.
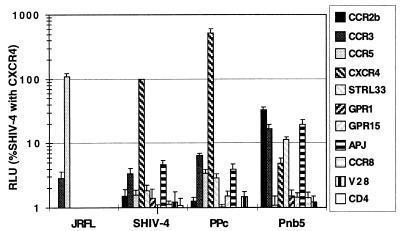
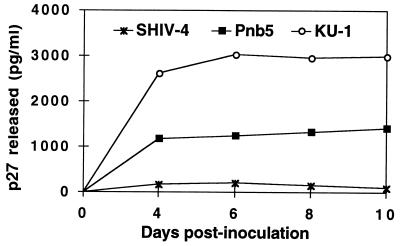
The Env protein from the parental SHIV-4 exclusively used CXCR4 as a coreceptor, as did multiple Env proteins derived from isolates obtained after serial passage through pig-tailed macaques (Fig. 1 and data not shown). However, one viral Env protein (Pnb5) obtained from virus isolated from the cerebral spinal fluid used CXCR4 to infect cells at levels that were only 4% of the parental virus. Far from being inactive, this Env protein efficiently used CCR2b as a coreceptor. In addition to CCR2b, the Pnb5 Env protein mediated infection of cells expressing either CCR3, STRL33, or APJ at levels considerably above background (>30-fold above background), though less efficiently than when CCR2b was present (Fig. 1). Results with rhesus CCR2b, CCR5, and CXCR4 were identical. Fusion was not observed when V28, CCR8, GPR15, or GPR1 were expressed in conjunction with CD4, even though sufficient levels of these coreceptors are expressed in our system to be used by other virus strains (27, 55). Despite the failure of the Pnb5 Env to use CCR5 and its relatively inefficient use of CXCR4, a replication competent SHIV containing the Pnb5 Env infected monocyte-derived macrophages from pig-tailed macaques, though not as efficiently as the well characterized macrophage-tropic SIV strain KU-1 (Fig. 2). Thus, changes in the Pnb5 Env relative to the parental SHIV sequence resulted in broadened coreceptor use and macrophage tropism.
Figure 1.
Coreceptor use profiles of SHIV Env proteins. Viral pseudotypes generated by cotransfection of env and NL-luc-E−R− constructs into 293T cells were used to infect feline CCCS target cells expressing CD4 and indicated coreceptor. Infection of target cells leads to integration of the reporter virus genome and long terminal repeat-mediated production of luciferase, which was quantified in cell lysates 3 days after infection. Values were normalized to the level of infection of SHIV-4 with CXCR4 for each experiment. Experiments were performed ≥3 times by using at least two independently derived virus stocks. Error bars represent SEMs. HIV-1 JRFL is shown as a control for CCR5 expression.
Figure 2.
The Pnb5 Env protein mediates infection of macaque macrophages. A recombinant virus containing the Pnb5 gp120 in a SHIV-4 background was constructed as described in Materials and Methods and used to inoculate rhesus macaque macrophage cultures. All cultures were inoculated with 1,000 TCID50 of each virus for 2 hr, washed three times to remove the virus inoculum, and maintained in macrophage differentiation medium for the course of the infection (44). Culture medium was harvested at the time points indicated and assayed for the presence of p27 antigen by using antigen capture assays.
Amino Acid Changes Associated with Altered Coreceptor Use.
Having identified a viral Env protein with a unique coreceptor use profile as a consequence of in vivo evolution, we sequenced the entire Pnb5 Env gene and compared it to the parental Env protein. The Pnb5 Env gp120 differed from the SHIV HXBc2 Env by 14 amino acids, nine of which were in regions corresponding to the variable loops of HIV-1 Env (Table 1). Five amino acid changes were present in the V3 loop, four of which affected basic residues. Because the acquisition of basic residues in the V3 loop is associated with CXCR4 use (10, 11, 21), substitution of some of these residues by neutral or acidic amino acids might be expected to result in the loss of CXCR4 reactivity. However, these changes did not enable the Env protein to use CCR5. Amino acid changes also were observed in V1/V2, V4, and V5. The K130N change in the stem of the V1/V2 loop introduces a potential glycosylation site, while T278M in the C3 region results in the loss of a carbohydrate addition site. The Pnb5 Env also had three conserved changes in gp41, none in functionally important regions (37). These did not contribute to altered coreceptor use or cytotropism.
Table 1.
Amino acid changes in Pnb5 Env
| AA position | Env region | AA change |
|---|---|---|
| 65 | C1 | V → A |
| 130 | C1 | K → N* |
| 192 | V2 | K → T |
| 202 | C2 | T → S |
| 270 | C2 | V → I |
| 278 | C2 | T → M† |
| 300 | V3 | N → S |
| 304 | V3 | R → I |
| 315 | V3 | R → G |
| 320 | V3 | I → M |
| 325 | V3 | N → D |
| 412 | V4 | D → G |
| 460 | V5 | N → K |
| 461 | V5 | S → G |
New-CHO site.
Lost-CHO site.
Generation and Analysis of Chimeric Env Proteins.
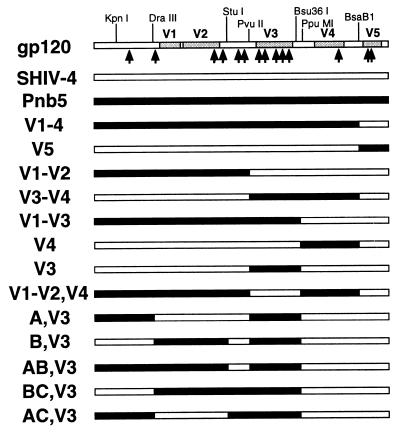
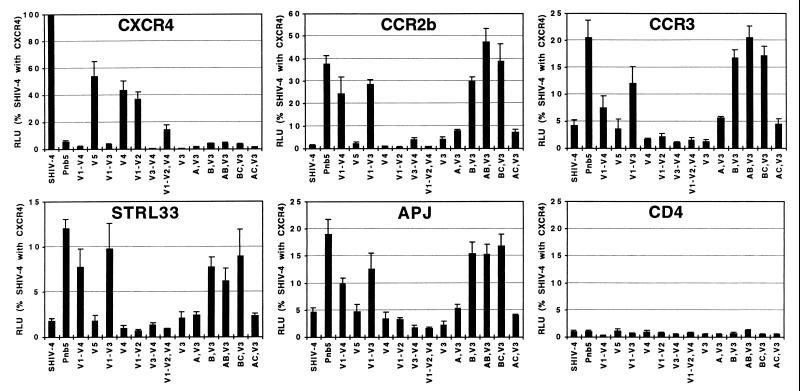
Chimeric Env proteins were generated to identify regions in the Pnb5 Env responsible for altered coreceptor use (Fig. 3). The resulting Envs were used to make pseudotype reporter viruses, which then were used to infect cells expressing CD4 and various coreceptors. We found that any construct containing the parental SHIV-4 V3 loop used CXCR4 to at least some extent. However, the efficiency of CXCR4 use was reduced by between one-half and two-thirds when the V1/2, V4, or V5 regions of Pnb5 were introduced into a SHIV-4 background. These effects were additive because introduction of both V1/2 and V4 regions of Pnb5 into SHIV-4 reduced CXCR4 use by approximately 10-fold. Conversely, any construct containing the Pnb5 V3 loop, either alone or in combination with other Pnb5 Env regions, was incapable of using CXCR4 as a coreceptor (Fig. 4). Taken together, these results demonstrate the central role of the V3 loop in governing use of CXCR4, with the five amino acid changes in this region between Pnb5 and SHIV-4 largely ablating CXCR4 use.
Figure 3.
Chimeric Env proteins. The gp120 diagram at the top indicates the restriction sites used to generate Env chimeras between SHIV-4 and Pnb5 and their positions relative to the variable regions of HIV-1 Env. Each arrow represents an amino acid difference between the SHIV-4 and Pnb5 Env proteins. The identity of these changes is listed in Table 1. Each chimera was tested for coreceptor use by pseudotype infection (Fig. 4) and for expression by Western blot (data not shown).
Figure 4.
Analysis of chimeric Env proteins. Pseudotype reporter viruses for each Env chimera shown in Fig. 3 was tested for infection against CCCS cells expressing CD4 and the indicated coreceptor. Values represent the average of ≥3 experiments using at least two independent virus stocks, and data are normalized to the level of infection of SHIV-4 with CXCR4 in each experiment. Error bars indicate SEMs.
Although introduction of the Pnb5 V3 loop into SHIV-4 largely abolished CXCR4 use, it was only sufficient to confer minimal activity with CCR2b or any other coreceptor. Likewise, the V1/2, V4, and V5 regions of Pnb5 failed to impart CCR2b use to the SHIV-4 Env protein when introduced singly. Rather, it was only the combination of V1/2 (six amino acid changes) in conjunction with V3 (five amino acid changes) that resulted in efficient CCR2b use. Thus, although the V3 loop is clearly an important determinant for CCR2b, it is not dominant, requiring changes in the V1/V2 region to impart activity with this alternative coreceptor. We also found that the Env determinants for use of CCR3, STRL33, and APJ were identical to those responsible for mediating infection of CCR2b-positive cells. Thus, inclusion of the Pnb5 V1/V2 and V3 loop regions conferred use of CCR2b, CCR3, STRL33, and APJ to the SHIV-4 Env while largely abolishing reactivity with CXCR4.
Analysis of Additional Env Chimeras.
A second series of Env chimeras were generated (Fig. 3) to more finely dissect the regions in the N-terminal half of the Pnb5 Env protein, which, in conjunction with changes in the V3 loop, resulted in altered coreceptor use. Analysis of these chimeric Env proteins indicated that the three amino acid changes in or near the V1/V2 region (K130N, K192T, and T202S) were responsible for altered coreceptor use, whereas changes in the C1 and C2 regions (V65A, V270I, and T278M) had no effect on coreceptor interactions (Fig. 4).
Env Expression and Processing.
Given the close relationship between the SHIV-4 and Pnb5 Env proteins, we anticipated that the chimeric Envs would be expressed and processed normally. However, because several chimeras failed to support infection under any circumstances, we analyzed the expression and processing of each Env protein. Media and cell lysates from 293T cells used to make viral pseudotypes were analyzed for Env by SDS/PAGE and Western blot by using a polyclonal rabbit serum generated against IIIB Env protein. We found that all chimeric Env proteins were processed in a similar fashion as judged by generation of gp120 from gp160 (data not shown). Thus, the failure of some Env chimeras (such as the Pnb5 V3 loop in a SHIV-4 background) to support infection of cells expressing either CXCR4 or CCR2b is likely caused by the fact that these Env proteins, despite normal processing, lack the necessary determinants to productively interact with any of the coreceptors tested.
DISCUSSION
During the course of HIV and SIV infection in humans and nonhuman primates, shifts in viral tropism associated with altered coreceptor use can occur that strongly influence viral pathogenesis and may impact disease course. Thus, emergence of X4 viruses has been correlated with accelerated disease progression (45, 46), while use of CCR3 has been linked to neuropathogenesis (34). The chemokine receptors also represent attractive targets for antiretroviral therapies because CCR5-negative individuals are highly resistant to virus infection (47–50). Furthermore, small molecule inhibitors targeted against CXCR4 have been described that block utilization of this coreceptor by HIV-1 strains (51–54). Therefore, identifying domains as well as specific residues in both Env and the coreceptors involved in virus entry will be important for understanding shifts in viral tropism, for the development of receptor antagonists, and for understanding the selective pressures that drive virus evolution in vivo. We have chosen to study the evolution of coreceptor use in vivo by examining virus isolates obtained from macaques infected with pathogenic and nonpathogenic SHIVs. By monitoring evolution in vivo, features may be recognized that are not obvious when constructing Env chimeras between two divergent Envs that exhibit different cytotropisms.
SHIVs based on X4 HIV-1 Env proteins have been shown to replicate in several nonhuman primates without causing disease (35). After serial passage of virus between monkeys, the virus undergoes an alteration in cytotropism and acquires the ability to infect macrophages and cause disease (36, 37). With the discovery of the coreceptors a potential explanation for this finding was provided: all SIV strains described to date, regardless of tropism, use CCR5 as a coreceptor (30–32, 38–40). CXCR4 is either not used by SIV or is used only rarely. Thus, introduction of a virus that exclusively uses CXCR4 into a nonhuman primate might be expected to result in either a virus that fails to replicate or in the rapid selection for virus strains that can use either CCR5 or other SIV coreceptors.
To our surprise, multiple SHIV isolates continued to exclusively use CXCR4 as a coreceptor. Although none of the SHIV Envs tested evolved to use CCR5 as a coreceptor, one Env clone (Pnb5) obtained from a virus swarm isolated from the cerebrospinal fluid (CSF) exhibited broadened coreceptor use as well as macrophage tropism. Interestingly, the Pnb5 Env did not use CCR5 and used CXCR4 inefficiently to infect cells, and thus represents a functional virus that does not use the major HIV-1 coreceptors. Instead, the Pnb5 Env protein used CCR2b, CCR3, STRL33, and APJ to infect cells, demonstrating that X4 viruses may evolve to use to other coreceptors in vivo. Of the four coreceptors used by the Pnb5 Env protein, CCR3 has been shown to function as a coreceptor for an impressive number of virus isolates, while STRL33 is commonly used by SIV strains to infect cells (24, 31, 55). Expression of these coreceptors on human primary T cells (CCR2b, CCR3, and STRL33), macrophages (CCR2b and STRL33), and transformed T-cell lines (APJ) is consistent with the possibility that any or all of these coreceptors could be used by this Env for infection in vivo. Furthermore, expression of CCR3 and APJ in the central nervous system may be relevant for replication of viruses bearing this CSF-derived Env in this compartment (30, 34).
Generation of chimeric Env proteins between SHIV-4 and Pnb5 indicated that the V3 loop is the major determinant of CXCR4 use. All chimeric Env proteins containing the SHIV-4 V3 loop used CXCR4 to at least some degree, though the efficiency with which CXCR4 could be used to infect cells depended in part on other regions of the Env protein. Thus, substitution of either the V1/2, V4, or V5 regions of SHIV-4 Env with the corresponding regions from Pnb5 resulted in less efficient use of the CXCR4 coreceptor. This finding is consistent with earlier studies that have shown that deletions or substitutions in V1/2 can influence the efficiency with which T-tropic viruses can infect cells (14–18). The Pnb5 V3 loop, which abolished CXCR4 utilization when introduced into the SHIV-4 background, contains two fewer basic residues than does the SHIV-4 V3 loop. Increased basic charge in the V3 region has been linked to T tropism as well as CXCR4 use, perhaps because the extracellular domains of CXCR4 most important for coreceptor activity are more acidic than the corresponding region in CCR5 (9–13).
Although the V3 loop of the SHIV-4 Env protein was both necessary and sufficient for CXCR4 use, the V3 loop of the Pnb5 Env protein did not confer efficient use of any coreceptor when placed in the SHIV-4 background. Rather, both the Pnb5 V3 loop and the V1/2 region in combination were necessary and sufficient for use of CCR2b, CCR3, STRL33, and APJ. These results indicate that the V1/2 region, although influencing use of CXCR4 and CCR5, plays a more important role in governing interactions with alternative coreceptors. Further, these results suggest that there is a common coreceptor binding site, because the determinants required to interact with multiple receptors appear to be similar. In addition, it is likely that these receptors possess conserved structural motifs because they are recognized by a single Env protein. Our results are consistent with a model in which the variable V1/2 and V3 regions interact with the coreceptors, at least initially. However, their variable nature and the fact that the determinants required for use of CCR2b, CCR3, STRL33, and APJ are similar also suggests that more conserved regions of Env are likely to make important interactions. The recently solved crystal structure of HIV-1 gp120 reveals a conserved coreceptor binding site adjacent to the V3 loop and also involving the stem of the V1/V2 region (Fig. 5; ref 56). Because the conserved regions of the SHIV-4 and Pnb5 Env proteins are essentially identical, we favor a model in which there are cooperative interactions between the V1/2 and V3 regions with a conserved coreceptor binding site in Env. The V3 loop may play a more important role in governing interactions with coreceptors that contain strongly negatively charged extracellular domains, such as CXCR4. Whether the V1/2 region contacts the coreceptor directly or in some way influences the conformation or accessibility of a conserved coreceptor binding site remains to be determined.
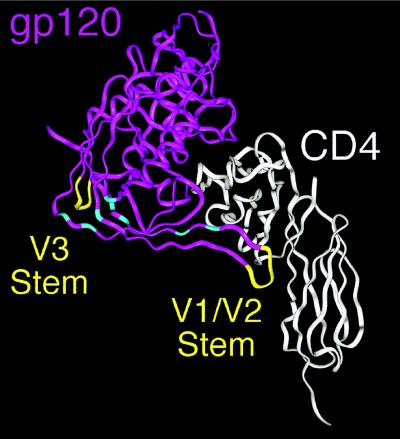
Figure 5.
Env determinants for coreceptor use. The ribbon diagram shows gp120 in purple and CD4 in white (56). The V1/V2 and V3 loops are lacking in the structure because of their truncation in the crystallized protein, but the points at which each emanate are shown in yellow. The Env determinants for coreceptor use in this paper reside in or very near to the V1/V2 and V3 loops. Conserved residues in gp120 shown by Rizzuto et al. (56) to play important roles in CCR5 binding are shown in light blue and reside in the bridging sheet, an antiparallel, four-stranded β-sheet structure that links the inner and outer domains of gp120 (56).
In addition to providing information on Env determinants responsible for coreceptor interactions, viruses such as Pnb5 with unique coreceptor use profiles can be used as probes to determine whether alternative coreceptors such as CCR2b, CCR3, STRL33, and APJ are expressed at levels and in a context that supports virus infection in both cell lines and primary cell populations. In addition, these viruses can be used to determine whether use of these receptors can lead to a productive infection in vivo as well as disease development. Finally, conserved sites in Env that mediate interactions with multiple coreceptors may be important targets for neutralizing antibodies as well as small molecule inhibitors.
Acknowledgments
We thank the members of the Doms lab for support and technical advice. A number of reagents used in these experiments were provided by the National Institutes of Health AIDS Reference and Reagent Program. This work was supported by National Institutes of Health Grants R01 AI-40880 (R.W.D.), DK49516 (E.B.S.), and AI38492, RR06753, and NS32203 (O.N.). T.L.H. was supported by the Franklin Scholars program.
ABBREVIATIONS
- Env
envelope
- SHIV
simian HIV
- HIV-1
HIV type 1
- SIV
simian immunodeficiency virus
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Doms R W, Peiper S C. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 2.Broder C C, Collman R G. J Leukocyte Biol. 1997;62:20–29. doi: 10.1002/jlb.62.1.20. [DOI] [PubMed] [Google Scholar]
- 3.Moore J P, Trkola A, Dragic T. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz P D, Cullen B R. Front Biosci. 1998;3:44–58. doi: 10.2741/a265. [DOI] [PubMed] [Google Scholar]
- 5.Berger E A. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 6.Schuitemaker H, Koot M, Koostra N A, Dercksen M W, Goede R E Y d, Steenwijk R P v, Lange J M A, Schattenkerk J K M E, Miedema F, Tersmette M. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Åsjö B, Morfeldt M L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo E M. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 8.Connor R I, Mohri H, Cao Y, Ho D D. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeJong J-J, Goudsmit J, Keulen W, Klaver B, Krone W, Tersmette M, deRonde A. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shioda T, Levy J A, Cheng-Mayer C. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Z, Berson J F, Chen Y, Turner J D, Zhang T, Sharron M, Jenks M H, Wang Z, Kim J, Rucker J, et al. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. J Virol. 1997;71:4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R W. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groenink M, Fouchier R A M, Broersen S, Baker C H, Koot M, Wout A B v t, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 18.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 20.Chesebro B, Nishis J, Perryman S, Cann A, O’Brien W, Chen I, Wehrly K. J Virol. 1991;65:5782–5789. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Jong J, De Ronde A, Keulen W, Tersmette M, Goudsmit J. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 24.Doms R W, Moore J P. In: Human Retroviruses and AIDS 1997—Los Alamos National Laboratory: Theoretical Biology and Biophysics. Korber B, Foley B, Leitner T, Myers G, Hahn B, McCutchan F, Mellors J, Kuiken C, editors. Los Alamos, NM: Los Alamos Natl Lab.; 1998. , Part III, pp. 1–12. [Google Scholar]
- 25.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 26.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, et al. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 27.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Collman R G, Doranz B J, Parmentier M, Doms R W. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samson M, Edinger A L, Stordeur P, Rucker J, Verhasselt V, Sharron M, Govaerts C, Mollereau C, Vassart G, Doms R W, Parmentier M. Eur J Immunol. 1998;28:1689–1700. doi: 10.1002/(SICI)1521-4141(199805)28:05<1689::AID-IMMU1689>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 29.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edinger, A. L., Hoffman, T. L., Yi, Y., Sharron, M., Collman, R. G., Mitrovic, B., Faulds, D., Hesselgesser, J., Horuk, R. & Doms, R. W. (1998) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 31.Deng H, Unutmaz D, Kewalramani V N, Littman D R. Nature (London) 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 32.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, et al. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeves J D, McKnight A, Potempa S, Simmons G, Gray P W, Power C A, Wells T, Weiss R A, Talbot S J. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 34.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, et al. Nature (London) 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Lord C I, Haseltine W, Letvin N L, Sodroski J. J Acquired Immune Defic Syndr. 1992;5:639–646. [PubMed] [Google Scholar]
- 36.Joag S V, Li Z, Foresman L, Stephens E B, Zhao L-J, Adany I, Pinson D M, McClure H M, Narayan O. J Virol. 1996;70:3189–3197. doi: 10.1128/jvi.70.5.3189-3197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens E B, Mukherjee S, Sahni M, Zhuge W, Raghavan R, Singh D K, Leung K, Atkinson B, Li Z, Joag S V, et al. Virology. 1997;231:313–321. doi: 10.1006/viro.1997.8534. [DOI] [PubMed] [Google Scholar]
- 38.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z, Clements J E, Murphey-Corb M, et al. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard N, Gerard C, Sodroski J. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens E B, Mukherjee S, Liu Z Q, Sheffer D, Lamb-Wharton R, Leung K, Zhuge W, Joag S V, Li Z, Foresman L, et al. J Virol. 1998;72:5207–5214. doi: 10.1128/jvi.72.6.5207-5214.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen B K, Saksela K, Andino R, Baltimore D. J Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connor R I, Chen B K, Choe S, Landau N R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 44.Stephens E B, McClure H M, Narayan O. Virology. 1995;206:535–544. doi: 10.1016/s0042-6822(95)80070-0. [DOI] [PubMed] [Google Scholar]
- 45.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scarlatti G, Tresoldi E, Björndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, et al. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 47.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 48.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapoumèroulie C, Cogniaux J, Forceille C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 49.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, et al. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 50.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, et al. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 51.Donzella G A, Schols D, Lin S W, Esté J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clerq E, Moore J P. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 52.Doranz B J, Grovit-Ferbas K, Sharron M P, Mao S-H, Goetz M B, Daar E S, Doms R W, O’Brien W A. J Exp Med. 1997;186:1395–1400. doi: 10.1084/jem.186.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, et al. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schols D, Struyf S, Van Damme J, Esté J A, Henson G, De Clerq E. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edinger, A. L., Hoffman, T. L., Sharron, M., Lee, B., O’Dowd, B. & Doms, R. W. (1998) Virology, in press. [DOI] [PubMed]
- 56.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]