Abstract
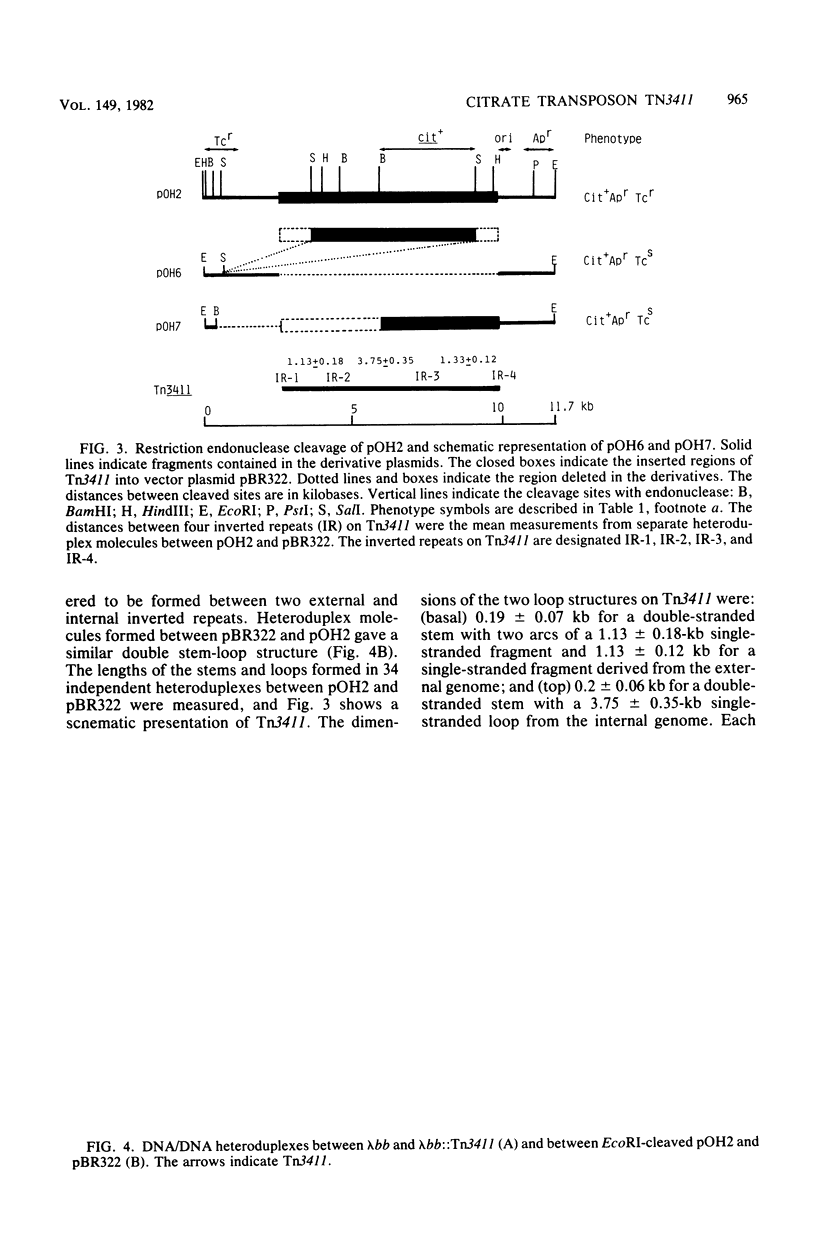
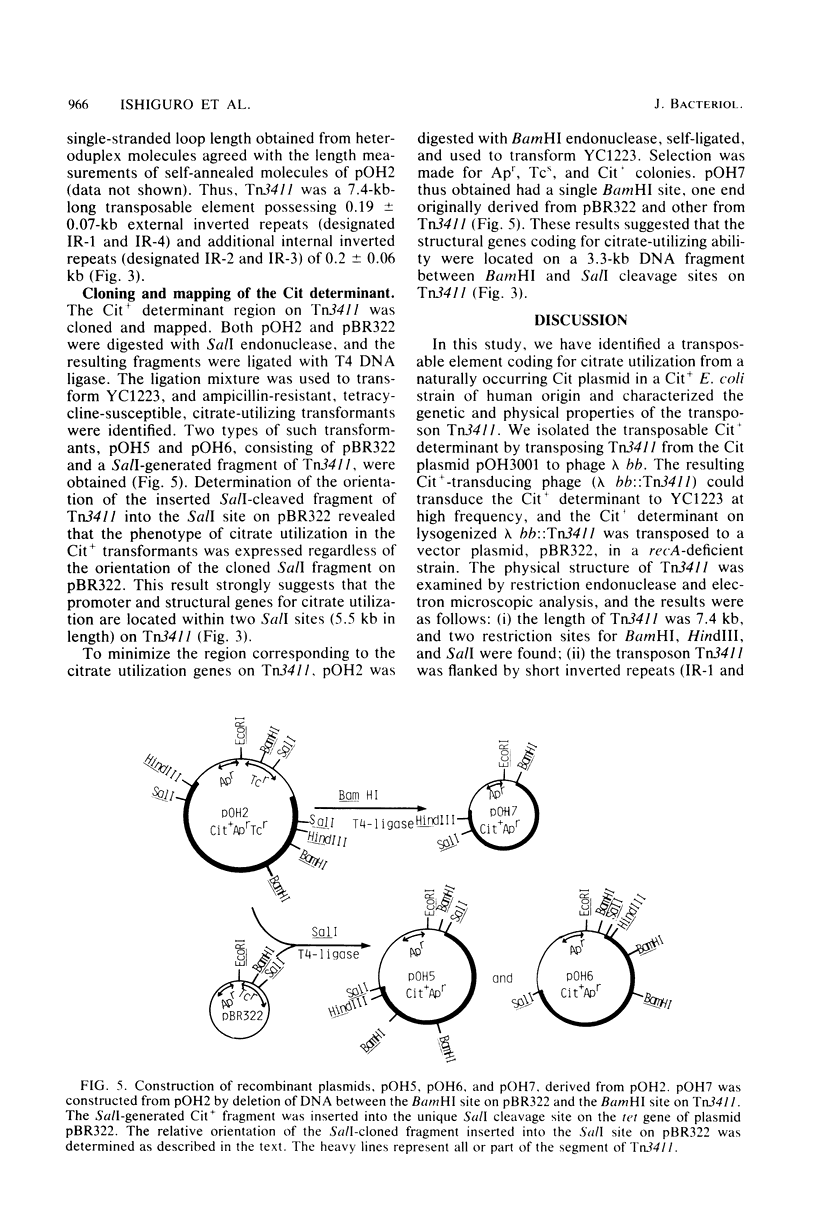
We have isolated a new transposon, Tn3411, encoding citrate-utilizing ability, from a naturally occurring citrate utilization (Cit) plasmid, pOH3001. Citrate transposon Tn3411 was transposed from pOH3001 to lambda b519 b515 cI857 S7 (abbreviated lambda bb) phage, and further from the resulting lambda bb:Tn3411 to a vector plasmid, pBR322, in recA-deficient strains. The Cit+ plasmids (pOH2 and pOH3) constructed by the integration of Tn3411 into pBR322 were examined by restriction endonuclease and heteroduplex analysis. The results obtained were as follows: (i) Tn3411 was 7.4 kilobases long and flanked by small inverted repeats, and it contained one more pair of inverted repeats at the opposite orientation in the internal region, thus making alternate repeats; and (ii) the Cit+ structure gene was located on the fragment (5.5 kilobases) between two SalI cleavage sites on Tn3411.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G., Ghosal D., Saedler H. Tn951: a new transposon carrying a lactose operon. Mol Gen Genet. 1978 Apr 6;160(2):215–224. doi: 10.1007/BF00267484. [DOI] [PubMed] [Google Scholar]
- Danbara H., Timmis J. K., Lurz R., Timmis K. N. Plasmid replication functions: two distinct segments of plasmid R1, RepA and RepD, express incompatibility and are capable of autonomous replication. J Bacteriol. 1980 Dec;144(3):1126–1138. doi: 10.1128/jb.144.3.1126-1138.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Ishiguro N., Hirose K., Asagi M., Sato G. Incompatibility of citrate utilization plasmids isolated from Escherichia coli. J Gen Microbiol. 1981 Mar;123(1):193–196. doi: 10.1099/00221287-123-1-193. [DOI] [PubMed] [Google Scholar]
- Ishiguro N., Hirose K., Sato G. Distribution of citrate utilization plasmids in Salmonella strains of bovine origin in Japan. Appl Environ Microbiol. 1980 Sep;40(3):446–451. doi: 10.1128/aem.40.3.446-451.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro N., Oka C., Hanzawa Y., Sato G. Isolation of citrate utilization plasmid from a bovine Salmonella typhimurium strain. Microbiol Immunol. 1980;24(8):757–760. doi: 10.1111/j.1348-0421.1980.tb02878.x. [DOI] [PubMed] [Google Scholar]
- Ishiguro N., Sato G. The distribution of plasmids determining citrate utilization in citrate-positive variants of Escherichia coli from humans, domestic animals, feral birds and environments. J Hyg (Lond) 1979 Oct;83(2):331–344. doi: 10.1017/s0022172400026127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupersztoch Y. M., Helinski D. R. A catenated DNA molecule as an intermediate in the replication of the resistance transfer factor R6K in Escherichia coli. Biochem Biophys Res Commun. 1973 Oct 15;54(4):1451–1459. doi: 10.1016/0006-291x(73)91149-2. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Smith H. W., Parsell Z., Green P. Thermosensitive H1 plasmids determining citrate utilization. J Gen Microbiol. 1978 Dec;109(2):305–311. doi: 10.1099/00221287-109-2-305. [DOI] [PubMed] [Google Scholar]
- So M., Heffron F., McCarthy B. J. The E. coli gene encoding heat stable toxin is a bacterial transposon flanked by inverted repeats of IS1. Nature. 1979 Feb 8;277(5696):453–456. doi: 10.1038/277453a0. [DOI] [PubMed] [Google Scholar]
- Starlinger P. IS elements and transposons. Plasmid. 1980 May;3(3):241–259. doi: 10.1016/0147-619x(80)90039-6. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Szybalski E. H., Szybalski W. A comprehensive molecular map of bacteriophage lambda. Gene. 1979 Nov;7(3-4):217–270. doi: 10.1016/0378-1119(79)90047-7. [DOI] [PubMed] [Google Scholar]
- Tu C. P., Cohen S. N. Translocation specificity of the Tn3 element: characterization of sites of multiple insertions. Cell. 1980 Jan;19(1):151–160. doi: 10.1016/0092-8674(80)90396-7. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R., Smith Grillo D., Isberg R., Way J., Syvanen M. Transposition of the kanamycin-resistance transposon Tn903. Mol Gen Genet. 1980;178(3):681–689. doi: 10.1007/BF00337879. [DOI] [PubMed] [Google Scholar]